Structural basis of GABAB receptor-Gi protein coupling
- PMID: 33911284
- PMCID: PMC8222003
- DOI: 10.1038/s41586-021-03507-1
Structural basis of GABAB receptor-Gi protein coupling
Abstract
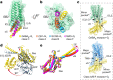
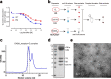
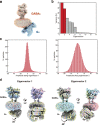
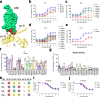
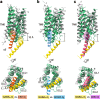
G-protein-coupled receptors (GPCRs) have central roles in intercellular communication1,2. Structural studies have revealed how GPCRs can activate G proteins. However, whether this mechanism is conserved among all classes of GPCR remains unknown. Here we report the structure of the class-C heterodimeric GABAB receptor, which is activated by the inhibitory transmitter GABA, in its active form complexed with Gi1 protein. We found that a single G protein interacts with the GB2 subunit of the GABAB receptor at a site that mainly involves intracellular loop 2 on the side of the transmembrane domain. This is in contrast to the G protein binding in a central cavity, as has been observed with other classes of GPCR. This binding mode results from the active form of the transmembrane domain of this GABAB receptor being different from that of other GPCRs, as it shows no outside movement of transmembrane helix 6. Our work also provides details of the inter- and intra-subunit changes that link agonist binding to G-protein activation in this heterodimeric complex.
Conflict of interest statement
The authors declare no competing interests.
Figures
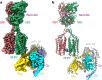







References
-
- Gilman AG. G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 1987;56:615–649. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

