Meningeal lymphatics affect microglia responses and anti-Aβ immunotherapy
- PMID: 33911285
- PMCID: PMC8817786
- DOI: 10.1038/s41586-021-03489-0
Meningeal lymphatics affect microglia responses and anti-Aβ immunotherapy
Abstract
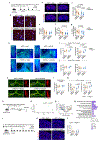
Alzheimer's disease (AD) is the most prevalent cause of dementia1. Although there is no effective treatment for AD, passive immunotherapy with monoclonal antibodies against amyloid beta (Aβ) is a promising therapeutic strategy2,3. Meningeal lymphatic drainage has an important role in the accumulation of Aβ in the brain4, but it is not known whether modulation of meningeal lymphatic function can influence the outcome of immunotherapy in AD. Here we show that ablation of meningeal lymphatic vessels in 5xFAD mice (a mouse model of amyloid deposition that expresses five mutations found in familial AD) worsened the outcome of mice treated with anti-Aβ passive immunotherapy by exacerbating the deposition of Aβ, microgliosis, neurovascular dysfunction, and behavioural deficits. By contrast, therapeutic delivery of vascular endothelial growth factor C improved clearance of Aβ by monoclonal antibodies. Notably, there was a substantial overlap between the gene signature of microglia from 5xFAD mice with impaired meningeal lymphatic function and the transcriptional profile of activated microglia from the brains of individuals with AD. Overall, our data demonstrate that impaired meningeal lymphatic drainage exacerbates the microglial inflammatory response in AD and that enhancement of meningeal lymphatic function combined with immunotherapies could lead to better clinical outcomes.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG057777/AG/NIA NIH HHS/United States
- AG034113/NH/NIH HHS/United States
- UF1 AG032438/AG/NIA NIH HHS/United States
- RF1 AG044546/AG/NIA NIH HHS/United States
- R01 NS118146/NS/NINDS NIH HHS/United States
- AG057496/NH/NIH HHS/United States
- RF1 AG057496/AG/NIA NIH HHS/United States
- U19 AG032438/AG/NIA NIH HHS/United States
- R56 AG067764/AG/NIA NIH HHS/United States
- P30 AG066444/AG/NIA NIH HHS/United States
- R01 AG062734/AG/NIA NIH HHS/United States
- U01 AG032438/AG/NIA NIH HHS/United States
- R37 AG034113/AG/NIA NIH HHS/United States
- RF1 AG053303/AG/NIA NIH HHS/United States
- R01 AG034113/AG/NIA NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- P50 AG005681/AG/NIA NIH HHS/United States
- P01 AG026276/AG/NIA NIH HHS/United States
- RF1 AG058501/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

