Reactivity of human antisera to codon optimized SARS-CoV2 viral proteins expressed in Escherichia coli
- PMID: 33912411
- PMCID: PMC8059472
- DOI: 10.4103/tcmj.tcmj_189_20
Reactivity of human antisera to codon optimized SARS-CoV2 viral proteins expressed in Escherichia coli
Abstract
Objective: The coronavirus disease 2019 (COVID-19) pandemic caused by the SARS-CoV2 virus continues to pose a serious threat to public health worldwide. The development of rapid diagnostic kits can assist the Tzu Chi Foundation in supporting global volunteers working to provide relief during the current pandemic.
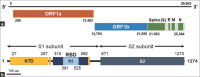
Materials and methods: In this study, nucleotide sequences derived from publicly available viral genome data for several domains of the SARS-CoV2 spike and nucleocapsid (N) proteins were chemically synthesized, with codon optimization for Escherichia coli protein expression. No actual viral particles were involved in these experiments. The synthesized sequences were cloned into an E. coli expression system based on pQE80L, and expressed viral proteins were subsequently purified using Ni-affinity chromatography. Western blotting was conducted using human antiviral sera to assess the response of codon-modified viral proteins to COVID-19 patient sera.
Results: N protein was expressed in amounts large enough to support large-scale production. The N-terminal domain, receptor-binding domain (RBD), Region 3, and the S2 domain were expressed in small but sufficient amounts for experiments. Immunoblotting results showed that anti-N IgG and anti-N IgM antibodies were detected in most patient sera, but only 60% of samples reacted with the recombinant RBD and S2 domain expressed by E. coli.
Conclusion: The results indicated that codon-optimized SARS-CoV2 viral proteins can be expressed in E. coli and purified for rapid antibody detection kit preparation, with the codon-optimized N protein, RBD, and S2 protein demonstrating the most potential.
Keywords: COVID-19; Codon optimization; Escherichia coli expression system; N protein; SARS-CoV2 antisera.
Copyright: © 2021 Tzu Chi Medical Journal.
Conflict of interest statement
There are no conflicts of interest.
Figures



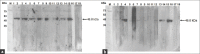

Similar articles
-
An efficient system to generate truncated human angiotensin converting enzyme 2 (hACE2) capable of binding RBD and spike protein of SARS-CoV2.Protein Expr Purif. 2021 Aug;184:105889. doi: 10.1016/j.pep.2021.105889. Epub 2021 Apr 11. Protein Expr Purif. 2021. PMID: 33852951 Free PMC article.
-
[Prokaryotic expression and characterization of two recombinant receptor-binding domain(RBD) proteins of human coronavirus NL63(HcoV-NL63)].Bing Du Xue Bao. 2013 Mar;29(2):106-11. Bing Du Xue Bao. 2013. PMID: 23757838 Chinese.
-
A Multi-Disulfide Receptor-Binding Domain (RBD) of the SARS-CoV-2 Spike Protein Expressed in E. coli Using a SEP-Tag Produces Antisera Interacting with the Mammalian Cell Expressed Spike (S1) Protein.Int J Mol Sci. 2022 Feb 1;23(3):1703. doi: 10.3390/ijms23031703. Int J Mol Sci. 2022. PMID: 35163624 Free PMC article.
-
Identification of immunodominant epitopes on nucleocapsid and spike proteins of the SARS-CoV-2 in Iranian COVID-19 patients.Pathog Dis. 2022 Feb 9;80(1):ftac001. doi: 10.1093/femspd/ftac001. Pathog Dis. 2022. PMID: 34994386 Free PMC article.
-
Heterogeneous antibodies against SARS-CoV-2 spike receptor binding domain and nucleocapsid with implications for COVID-19 immunity.JCI Insight. 2020 Sep 17;5(18):e142386. doi: 10.1172/jci.insight.142386. JCI Insight. 2020. PMID: 32796155 Free PMC article.
References
-
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–20. - PubMed
-
- Nguyen NL, Kim JM, Park JA, Park SM, Jang YS, Yang MS, et al. Expression and purification of an immunogenic dengue virus epitope using a synthetic consensus sequence of envelope domain III and Saccharomyces cerevisiae. Protein Expr Purif. 2013;88:235–42. - PubMed
-
- Nguyen NL, So KK, Kim JM, Kim SH, Jang YS, Yang MS, et al. Expression and characterization of an M cell-specific ligand-fused dengue virus tetravalent epitope using Saccharomyces cerevisiae. J Biosci Bioeng. 2015;119:19–27. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
