A molecular single-cell lung atlas of lethal COVID-19
- PMID: 33915568
- PMCID: PMC8814825
- DOI: 10.1038/s41586-021-03569-1
A molecular single-cell lung atlas of lethal COVID-19
Erratum in
-
Author Correction: A molecular single-cell lung atlas of lethal COVID-19.Nature. 2021 Oct;598(7882):E2. doi: 10.1038/s41586-021-03921-5. Nature. 2021. PMID: 34625743 Free PMC article. No abstract available.
Abstract
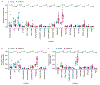
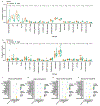
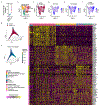
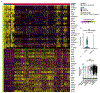
Respiratory failure is the leading cause of death in patients with severe SARS-CoV-2 infection1,2, but the host response at the lung tissue level is poorly understood. Here we performed single-nucleus RNA sequencing of about 116,000 nuclei from the lungs of nineteen individuals who died of COVID-19 and underwent rapid autopsy and seven control individuals. Integrated analyses identified substantial alterations in cellular composition, transcriptional cell states, and cell-to-cell interactions, thereby providing insight into the biology of lethal COVID-19. The lungs from individuals with COVID-19 were highly inflamed, with dense infiltration of aberrantly activated monocyte-derived macrophages and alveolar macrophages, but had impaired T cell responses. Monocyte/macrophage-derived interleukin-1β and epithelial cell-derived interleukin-6 were unique features of SARS-CoV-2 infection compared to other viral and bacterial causes of pneumonia. Alveolar type 2 cells adopted an inflammation-associated transient progenitor cell state and failed to undergo full transition into alveolar type 1 cells, resulting in impaired lung regeneration. Furthermore, we identified expansion of recently described CTHRC1+ pathological fibroblasts3 contributing to rapidly ensuing pulmonary fibrosis in COVID-19. Inference of protein activity and ligand-receptor interactions identified putative drug targets to disrupt deleterious circuits. This atlas enables the dissection of lethal COVID-19, may inform our understanding of long-term complications of COVID-19 survivors, and provides an important resource for therapeutic development.
Conflict of interest statement
Figures









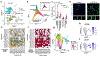

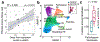
Comment in
-
Single-cell atlases dissect tissue destruction by SARS-CoV-2.Nat Rev Immunol. 2021 Jun;21(6):343. doi: 10.1038/s41577-021-00564-5. Nat Rev Immunol. 2021. PMID: 33990788 Free PMC article.
-
COVID-19 lung atlas.Nat Cell Biol. 2021 Nov;23(11):1108. doi: 10.1038/s41556-021-00779-7. Nat Cell Biol. 2021. PMID: 34750573 No abstract available.
References
-
- Dong Y et al. Epidemiology of COVID-19 among children in china. Pediatrics 145, e20200702 (2020). - PubMed
-
- Bellani G et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. J. Am. Med. Assoc. 315, 788–800 (2016). - PubMed
Publication types
MeSH terms
Grants and funding
- T32 CA203702/CA/NCI NIH HHS/United States
- K08 CA222663/CA/NCI NIH HHS/United States
- R56 AR078686/AR/NIAMS NIH HHS/United States
- R01 CA194547/CA/NCI NIH HHS/United States
- R01 CA234614/CA/NCI NIH HHS/United States
- UL1 TR002384/TR/NCATS NIH HHS/United States
- R01 AI107301/AI/NIAID NIH HHS/United States
- R21 AI163753/AI/NIAID NIH HHS/United States
- R37 CA258829/CA/NCI NIH HHS/United States
- U54 CA225088/CA/NCI NIH HHS/United States
- R01 HL152293/HL/NHLBI NIH HHS/United States
- S10 RR027050/RR/NCRR NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- T32 AI106711/AI/NIAID NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 HL132996/HL/NHLBI NIH HHS/United States
- P30 CA013696/CA/NCI NIH HHS/United States
- R03 DK117252/DK/NIDDK NIH HHS/United States
- R01 HL159675/HL/NHLBI NIH HHS/United States
- R01 DK121072/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

