Nosocomial Outbreak of SARS-CoV-2 in a "Non-COVID-19" Hospital Ward: Virus Genome Sequencing as a Key Tool to Understand Cryptic Transmission
- PMID: 33916205
- PMCID: PMC8065743
- DOI: 10.3390/v13040604
Nosocomial Outbreak of SARS-CoV-2 in a "Non-COVID-19" Hospital Ward: Virus Genome Sequencing as a Key Tool to Understand Cryptic Transmission
Abstract
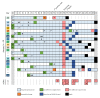
Dissemination of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in healthcare institutions affects both patients and health-care workers (HCW), as well as the institutional capacity to provide essential health services. Here, we investigated an outbreak of SARS-CoV-2 in a "non-COVID-19" hospital ward unveiled by massive testing, which challenged the reconstruction of transmission chains. The contacts network during the 15-day period before the screening was investigated, and positive SARS-CoV-2 RNA samples were subjected to virus genome sequencing. Of the 245 tested individuals, 48 (21 patients and 27 HCWs) tested positive for SARS-CoV-2. HCWs were mostly asymptomatic, but the mortality among patients reached 57.1% (12/21). Phylogenetic reconstruction revealed that all cases were part of the same transmission chain. By combining contact tracing and genomic data, including analysis of emerging minor variants, we unveiled a scenario of silent SARS-CoV-2 dissemination, mostly driven by the close contact within the HCWs group and between HCWs and patients. This investigation triggered enhanced prevention and control measures, leading to more timely detection and containment of novel outbreaks. This study shows the benefit of combining genomic and epidemiological data for disclosing complex nosocomial outbreaks, and provides valuable data to prevent transmission of COVID-19 in healthcare facilities.
Keywords: COVID-19; SARS-CoV-2; contact tracing; genome sequencing; healthcare institution; nosocomial outbreak.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- World Health Organization (WHO) Coronavirus Disease (COVID-19) Weekly Epidemiological Update 8 December 2020. [(accessed on 13 December 2020)]; Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio....
-
- World Health Organization (WHO) Infection Prevention and Control During Health Care When Coronavirus Disease (COVID-19) is Suspected or Confirmed. Geneva: World Health Organization. [(accessed on 11 October 2020)];2020 Available online: https://apps.who.int/iris/rest/bitstreams/1284718/retrieve.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

