MicroRNA Expression Profile Distinguishes Glioblastoma Stem Cells from Differentiated Tumor Cells
- PMID: 33916317
- PMCID: PMC8066769
- DOI: 10.3390/jpm11040264
MicroRNA Expression Profile Distinguishes Glioblastoma Stem Cells from Differentiated Tumor Cells
Abstract
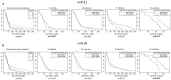
Glioblastoma (GBM) represents the most common and aggressive tumor of the brain. Despite the fact that several studies have recently addressed the molecular mechanisms underlying the disease, its etiology and pathogenesis are still poorly understood. GBM displays poor prognosis and its resistance to common therapeutic approaches makes it a highly recurrent tumor. Several studies have identified a subpopulation of tumor cells, known as GBM cancer stem cells (CSCs) characterized by the ability of self-renewal, tumor initiation and propagation. GBM CSCs have been shown to survive GBM chemotherapy and radiotherapy. Thus, targeting CSCs represents a promising approach to treat GBM. Recent evidence has shown that GBM is characterized by a dysregulated expression of microRNA (miRNAs). In this study we have investigated the difference between human GBM CSCs and their paired autologous differentiated tumor cells. Array-based profiling and quantitative Real-Time PCR (qRT-PCR) were performed to identify miRNAs differentially expressed in CSCs. The Cancer Genome Atlas (TCGA) data were also interrogated, and functional interpretation analysis was performed. We have identified 14 miRNAs significantly differentially expressed in GBM CSCs (p < 0.005). MiR-21 and miR-95 were among the most significantly deregulated miRNAs, and their expression was also associated to patient survival. We believe that the data provided here carry important implications for future studies aiming at elucidating the molecular mechanisms underlying GBM.
Keywords: cancer; cancer stem cells; glioblastoma; microRNAs; qPCR.
Conflict of interest statement
The authors declare no conflict of interest or relationships relevant to the content of this paper. All authors take responsibility for all aspects of the reliability of the data presented and their discussed interpretation.
Figures



References
-
- Luo J.W., Wang X., Yang Y., Mao Q. Role of micro-RNA (miRNA) in pathogenesis of glioblastoma. Eur. Rev. Med. Pharmacol. Sci. 2015;19:1630–1639. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

