Detecting and Characterizing A-To-I microRNA Editing in Cancer
- PMID: 33916692
- PMCID: PMC8038323
- DOI: 10.3390/cancers13071699
Detecting and Characterizing A-To-I microRNA Editing in Cancer
Abstract
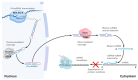
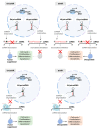
Adenosine to inosine (A-to-I) editing consists of an RNA modification where single adenosines along the RNA sequence are converted into inosines. Such a biochemical transformation is catalyzed by enzymes belonging to the family of adenosine deaminases acting on RNA (ADARs) and occurs either co- or post-transcriptionally. The employment of powerful, high-throughput detection methods has recently revealed that A-to-I editing widely occurs in non-coding RNAs, including microRNAs (miRNAs). MiRNAs are a class of small regulatory non-coding RNAs (ncRNAs) acting as translation inhibitors, known to exert relevant roles in controlling cell cycle, proliferation, and cancer development. Indeed, a growing number of recent researches have evidenced the importance of miRNA editing in cancer biology by exploiting various detection and validation methods. Herein, we briefly overview early and currently available A-to-I miRNA editing detection and validation methods and discuss the significance of A-to-I miRNA editing in human cancer.
Keywords: A-to-I RNA editing; ADAR; detection; functional characterization; microRNA targeting; microRNAs; quantification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Grosjean H. Fine-Tuning of RNA Functions by Modification and Editing. Springer; Berlin/Heidelberg, Germany: 2005. Modification and editing of RNA: Historical overview and important facts to remember; pp. 1–22.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

