FragNet, a Contrastive Learning-Based Transformer Model for Clustering, Interpreting, Visualizing, and Navigating Chemical Space
- PMID: 33916824
- PMCID: PMC8038408
- DOI: 10.3390/molecules26072065
FragNet, a Contrastive Learning-Based Transformer Model for Clustering, Interpreting, Visualizing, and Navigating Chemical Space
Abstract
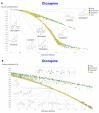
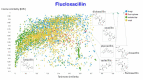
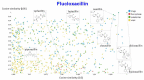
The question of molecular similarity is core in cheminformatics and is usually assessed via a pairwise comparison based on vectors of properties or molecular fingerprints. We recently exploited variational autoencoders to embed 6M molecules in a chemical space, such that their (Euclidean) distance within the latent space so formed could be assessed within the framework of the entire molecular set. However, the standard objective function used did not seek to manipulate the latent space so as to cluster the molecules based on any perceived similarity. Using a set of some 160,000 molecules of biological relevance, we here bring together three modern elements of deep learning to create a novel and disentangled latent space, viz transformers, contrastive learning, and an embedded autoencoder. The effective dimensionality of the latent space was varied such that clear separation of individual types of molecules could be observed within individual dimensions of the latent space. The capacity of the network was such that many dimensions were not populated at all. As before, we assessed the utility of the representation by comparing clozapine with its near neighbors, and we also did the same for various antibiotics related to flucloxacillin. Transformers, especially when as here coupled with contrastive learning, effectively provide one-shot learning and lead to a successful and disentangled representation of molecular latent spaces that at once uses the entire training set in their construction while allowing "similar" molecules to cluster together in an effective and interpretable way.
Keywords: artificial intelligence; attention; chemical space; cheminformatics; deep learning; generative methods; neural networks; transformers.
Conflict of interest statement
The authors declare that they have no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





















References
-
- Brown T.B., Mann B., Ryder N., Subbiah M., Kaplan J., Dhariwal P., Neelakantan A., Shyam P., Sastry G., Askell A., et al. Language models are few-shot learners. arXiv. 20202005.14165
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

