Human Mitoribosome Biogenesis and Its Emerging Links to Disease
- PMID: 33917098
- PMCID: PMC8067846
- DOI: 10.3390/ijms22083827
Human Mitoribosome Biogenesis and Its Emerging Links to Disease
Abstract
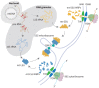
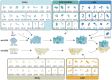
Mammalian mitochondrial ribosomes (mitoribosomes) synthesize a small subset of proteins, which are essential components of the oxidative phosphorylation machinery. Therefore, their function is of fundamental importance to cellular metabolism. The assembly of mitoribosomes is a complex process that progresses through numerous maturation and protein-binding events coordinated by the actions of several assembly factors. Dysregulation of mitoribosome production is increasingly recognized as a contributor to metabolic and neurodegenerative diseases. In recent years, mutations in multiple components of the mitoribosome assembly machinery have been associated with a range of human pathologies, highlighting their importance to cell function and health. Here, we provide a review of our current understanding of mitoribosome biogenesis, highlighting the key factors involved in this process and the growing number of mutations in genes encoding mitoribosomal RNAs, proteins, and assembly factors that lead to human disease.
Keywords: assembly factors; mitochondria; mitochondrial disease; mitoribosome; rRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The Diseased Mitoribosome.FEBS Lett. 2021 Apr;595(8):1025-1061. doi: 10.1002/1873-3468.14024. Epub 2020 Dec 22. FEBS Lett. 2021. PMID: 33314036 Free PMC article. Review.
-
A roadmap for ribosome assembly in human mitochondria.Nat Struct Mol Biol. 2024 Dec;31(12):1898-1908. doi: 10.1038/s41594-024-01356-w. Epub 2024 Jul 11. Nat Struct Mol Biol. 2024. PMID: 38992089 Free PMC article.
-
The human Obg protein GTPBP10 is involved in mitoribosomal biogenesis.Nucleic Acids Res. 2018 Sep 19;46(16):8471-8482. doi: 10.1093/nar/gky701. Nucleic Acids Res. 2018. PMID: 30085210 Free PMC article.
-
Mitoribosomal small subunit maturation involves formation of initiation-like complexes.Proc Natl Acad Sci U S A. 2022 Jan 18;119(3):e2114710118. doi: 10.1073/pnas.2114710118. Proc Natl Acad Sci U S A. 2022. PMID: 35042777 Free PMC article.
-
Role of GTPases in Driving Mitoribosome Assembly.Trends Cell Biol. 2021 Apr;31(4):284-297. doi: 10.1016/j.tcb.2020.12.008. Epub 2021 Jan 5. Trends Cell Biol. 2021. PMID: 33419649 Free PMC article. Review.
Cited by
-
Biallelic variants in DAP3 result in reduced assembly of the mitoribosomal small subunit with altered intrinsic and extrinsic apoptosis and a Perrault syndrome-spectrum phenotype.medRxiv [Preprint]. 2024 Aug 21:2024.08.19.24312079. doi: 10.1101/2024.08.19.24312079. medRxiv. 2024. Update in: Am J Hum Genet. 2025 Jan 2;112(1):59-74. doi: 10.1016/j.ajhg.2024.11.007. PMID: 39371131 Free PMC article. Updated. Preprint.
-
Mitochondrial Protein Translation: Emerging Roles and Clinical Significance in Disease.Front Cell Dev Biol. 2021 Jul 1;9:675465. doi: 10.3389/fcell.2021.675465. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34277617 Free PMC article. Review.
-
Four-Color STED Super-Resolution RNA Fluorescent In Situ Hybridization and Immunocytochemistry to Visualize Mitochondrial mRNAs in Context with Mitochondrial Ribosomes.Methods Mol Biol. 2023;2661:303-316. doi: 10.1007/978-1-0716-3171-3_17. Methods Mol Biol. 2023. PMID: 37166644
-
Mitochondrial ribosome biogenesis and redox sensing.FEBS Open Bio. 2024 Oct;14(10):1640-1655. doi: 10.1002/2211-5463.13844. Epub 2024 Jun 7. FEBS Open Bio. 2024. PMID: 38849194 Free PMC article. Review.
-
Altered mitochondrial microenvironment at the spotlight of musculoskeletal aging and Alzheimer's disease.Sci Rep. 2022 Jul 4;12(1):11290. doi: 10.1038/s41598-022-15578-9. Sci Rep. 2022. PMID: 35788655 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

