Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19
- PMID: 33917481
- PMCID: PMC8067447
- DOI: 10.3390/cells10040821
Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19
Abstract
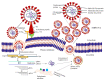
Coronavirus belongs to the family of Coronaviridae, comprising single-stranded, positive-sense RNA genome (+ ssRNA) of around 26 to 32 kilobases, and has been known to cause infection to a myriad of mammalian hosts, such as humans, cats, bats, civets, dogs, and camels with varied consequences in terms of death and debilitation. Strikingly, novel coronavirus (2019-nCoV), later renamed as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), and found to be the causative agent of coronavirus disease-19 (COVID-19), shows 88% of sequence identity with bat-SL-CoVZC45 and bat-SL-CoVZXC21, 79% with SARS-CoV and 50% with MERS-CoV, respectively. Despite key amino acid residual variability, there is an incredible structural similarity between the receptor binding domain (RBD) of spike protein (S) of SARS-CoV-2 and SARS-CoV. During infection, spike protein of SARS-CoV-2 compared to SARS-CoV displays 10-20 times greater affinity for its cognate host cell receptor, angiotensin-converting enzyme 2 (ACE2), leading proteolytic cleavage of S protein by transmembrane protease serine 2 (TMPRSS2). Following cellular entry, the ORF-1a and ORF-1ab, located downstream to 5' end of + ssRNA genome, undergo translation, thereby forming two large polyproteins, pp1a and pp1ab. These polyproteins, following protease-induced cleavage and molecular assembly, form functional viral RNA polymerase, also referred to as replicase. Thereafter, uninterrupted orchestrated replication-transcription molecular events lead to the synthesis of multiple nested sets of subgenomic mRNAs (sgRNAs), which are finally translated to several structural and accessory proteins participating in structure formation and various molecular functions of virus, respectively. These multiple structural proteins assemble and encapsulate genomic RNA (gRNA), resulting in numerous viral progenies, which eventually exit the host cell, and spread infection to rest of the body. In this review, we primarily focus on genomic organization, structural and non-structural protein components, and potential prospective molecular targets for development of therapeutic drugs, convalescent plasm therapy, and a myriad of potential vaccines to tackle SARS-CoV-2 infection.
Keywords: Angiotensin converting enzyme 2; Coronavirus disease-19; SARS-CoV-2; coronavirus; structural proteins.
Conflict of interest statement
Authors declare no conflict of interest for this article.
Figures

References
-
- Rodriguez-Morales A.J., Bonilla-Aldana D.K., Balbin-Ramon G.J., Rabaan A.A., Sah R., Paniz-Mondolfi A., Pagliano P., Esposito S. History is repeating itself: Probable zoonotic spillover as the cause of the 2019 novel Coronavirus Epidemic. Infez. Med. 2020;28:3–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

