Active Rumen Bacterial and Protozoal Communities Revealed by RNA-Based Amplicon Sequencing on Dairy Cows Fed Different Diets at Three Physiological Stages
- PMID: 33918504
- PMCID: PMC8066057
- DOI: 10.3390/microorganisms9040754
Active Rumen Bacterial and Protozoal Communities Revealed by RNA-Based Amplicon Sequencing on Dairy Cows Fed Different Diets at Three Physiological Stages
Abstract
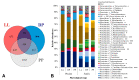
Seven Italian Simmental cows were monitored during three different physiological stages, namely late lactation (LL), dry period (DP), and postpartum (PP), to evaluate modifications in their metabolically-active rumen bacterial and protozoal communities using the RNA-based amplicon sequencing method. The bacterial community was dominated by seven phyla: Proteobacteria, Bacteroidetes, Firmicutes, Spirochaetes, Fibrobacteres, Verrucomicrobia, and Tenericutes. The relative abundance of the phylum Proteobacteria decreased from 47.60 to 28.15% from LL to DP and then increased to 33.24% in PP. An opposite pattern in LL, DP, and PP stages was observed for phyla Verrucomicrobia (from 0.96 to 4.30 to 1.69%), Elusimicrobia (from 0.32 to 2.84 to 0.25%), and SR1 (from 0.50 to 2.08 to 0.79%). The relative abundance of families Succinivibrionaceae and Prevotellaceae decreased in the DP, while Ruminococcaceae increased. Bacterial genera Prevotella and Treponema were least abundant in the DP as compared to LL and PP, while Ruminobacter and Succinimonas were most abundant in the DP. The rumen eukaryotic community was dominated by protozoal phylum Ciliophora, which showed a significant decrease in relative abundance from 97.6 to 93.9 to 92.6 in LL, DP, and PP, respectively. In conclusion, the physiological stage-dependent dietary changes resulted in a clear shift in metabolically-active rumen microbial communities.
Keywords: RNA; active rumen microbiota; cDNA; physiological stages; protozoa.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Community structure of the metabolically active rumen bacterial and archaeal communities of dairy cows over the transition period.PLoS One. 2017 Nov 8;12(11):e0187858. doi: 10.1371/journal.pone.0187858. eCollection 2017. PLoS One. 2017. PMID: 29117259 Free PMC article.
-
Factors influencing ruminal bacterial community diversity and composition and microbial fibrolytic enzyme abundance in lactating dairy cows with a focus on the role of active dry yeast.J Dairy Sci. 2017 Jun;100(6):4377-4393. doi: 10.3168/jds.2016-11473. Epub 2017 Apr 5. J Dairy Sci. 2017. PMID: 28390722 Clinical Trial.
-
Stability Assessment of the Rumen Bacterial and Archaeal Communities in Dairy Cows Within a Single Lactation and Its Association With Host Phenotype.Front Microbiol. 2021 Apr 6;12:636223. doi: 10.3389/fmicb.2021.636223. eCollection 2021. Front Microbiol. 2021. PMID: 33927700 Free PMC article.
-
Assessment of rumen bacteria in dairy cows with varied milk protein yield.J Dairy Sci. 2019 Jun;102(6):5031-5041. doi: 10.3168/jds.2018-15974. Epub 2019 Apr 10. J Dairy Sci. 2019. PMID: 30981485
-
Composition of bacterial and archaeal communities in the rumen of dromedary camel using cDNA-amplicon sequencing.Int Microbiol. 2020 May;23(2):137-148. doi: 10.1007/s10123-019-00093-1. Epub 2019 Aug 20. Int Microbiol. 2020. PMID: 31432356 Review.
Cited by
-
Transmission of fungi and protozoa under grazing conditions from lactating yaks to sucking yak calves in early life.Appl Microbiol Biotechnol. 2023 Aug;107(15):4931-4945. doi: 10.1007/s00253-023-12616-y. Epub 2023 Jun 21. Appl Microbiol Biotechnol. 2023. PMID: 37341753 Free PMC article.
-
Ruminal background of predisposed milk urea (MU) concentration in Holsteins.Front Microbiol. 2022 Sep 13;13:939711. doi: 10.3389/fmicb.2022.939711. eCollection 2022. Front Microbiol. 2022. PMID: 36177471 Free PMC article.
-
Bovine Animal Model for Studying the Maternal Microbiome, in utero Microbial Colonization and Their Role in Offspring Development and Fetal Programming.Front Microbiol. 2022 Feb 23;13:854453. doi: 10.3389/fmicb.2022.854453. eCollection 2022. Front Microbiol. 2022. PMID: 35283808 Free PMC article. Review.
-
Heat Stress Induces Shifts in the Rumen Bacteria and Metabolome of Buffalo.Animals (Basel). 2022 May 18;12(10):1300. doi: 10.3390/ani12101300. Animals (Basel). 2022. PMID: 35625146 Free PMC article.
-
Under heat stress conditions, selenium nanoparticles promote lactation through modulation of rumen microbiota and metabolic processes in dairy goats.Sci Rep. 2025 Mar 17;15(1):9063. doi: 10.1038/s41598-025-93710-1. Sci Rep. 2025. PMID: 40097638 Free PMC article.
References
-
- Petri R.M., Schwaiger T., Penner G.B., Beauchemin K.A., Forster R.J., McKinnon J.J., McAllister T.A. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE. 2013;8:e83424. doi: 10.1371/journal.pone.0083424. - DOI - PMC - PubMed
-
- Zened A., Combes S., Cauquil L., Mariette J., Klopp C., Bouchez O., Troegeler-Meynadier A., Enjalbert F. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol. Ecol. 2013;83:504–514. doi: 10.1111/1574-6941.12011. - DOI - PubMed
-
- Ghaffari M.H., Tahmasbi A.M., Khorvash M., Naserian A.A., Ghaffari A.H., Valizadeh H. Effects of pistachio by-products in replacement of alfalfa hay on populations of rumen bacteria involved in biohydrogenation and fermentative parameters in the rumen of sheep. J. Anim. Physiol. Anim. Nutr. 2014;98:578–586. doi: 10.1111/jpn.12120. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

