The Phylogeography of Potato Virus X Shows the Fingerprints of Its Human Vector
- PMID: 33918611
- PMCID: PMC8070401
- DOI: 10.3390/v13040644
The Phylogeography of Potato Virus X Shows the Fingerprints of Its Human Vector
Abstract
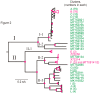
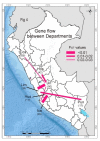
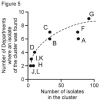
Potato virus X (PVX) occurs worldwide and causes an important potato disease. Complete PVX genomes were obtained from 326 new isolates from Peru, which is within the potato crop's main domestication center, 10 from historical PVX isolates from the Andes (Bolivia, Peru) or Europe (UK), and three from Africa (Burundi). Concatenated open reading frames (ORFs) from these genomes plus 49 published genomic sequences were analyzed. Only 18 of them were recombinants, 17 of them Peruvian. A phylogeny of the non-recombinant sequences found two major (I, II) and five minor (I-1, I-2, II-1, II-2, II-3) phylogroups, which included 12 statistically supported clusters. Analysis of 488 coat protein (CP) gene sequences, including 128 published previously, gave a completely congruent phylogeny. Among the minor phylogroups, I-2 and II-3 only contained Andean isolates, I-1 and II-2 were of both Andean and other isolates, but all of the three II-1 isolates were European. I-1, I-2, II-1 and II-2 all contained biologically typed isolates. Population genetic and dating analyses indicated that PVX emerged after potato's domestication 9000 years ago and was transported to Europe after the 15th century. Major clusters A-D probably resulted from expansions that occurred soon after the potato late-blight pandemic of the mid-19th century. Genetic comparisons of the PVX populations of different Peruvian Departments found similarities between those linked by local transport of seed potato tubers for summer rain-watered highland crops, and those linked to winter-irrigated crops in nearby coastal Departments. Comparisons also showed that, although the Andean PVX population was diverse and evolving neutrally, its spread to Europe and then elsewhere involved population expansion. PVX forms a basal Potexvirus genus lineage but its immediate progenitor is unknown. Establishing whether PVX's entirely Andean phylogroups I-2 and II-3 and its Andean recombinants threaten potato production elsewhere requires future biological studies.
Keywords: Andean crop domestication center; Andean lineages; South America; biosecurity significance; dating; evolution; high-throughput sequencing; interpretation; phylogenetics; population genetics; potato; potato virus X; prehistory; strain groups; virus disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Phylogenetics and Evolution of Potato Virus V: Another Potyvirus that Originated in the Andes.Plant Dis. 2022 Feb;106(2):691-700. doi: 10.1094/PDIS-09-21-1897-RE. Epub 2022 Feb 8. Plant Dis. 2022. PMID: 34633236
-
Potato Virus A Isolates from Three Continents: Their Biological Properties, Phylogenetics, and Prehistory.Phytopathology. 2021 Jan;111(1):217-226. doi: 10.1094/PHYTO-08-20-0354-FI. Epub 2020 Dec 15. Phytopathology. 2021. PMID: 33174824
-
Complete genome sequences of new divergent potato virus X isolates and discrimination between strains in a mixed infection using small RNAs sequencing approach.Virus Res. 2014 Oct 13;191:45-50. doi: 10.1016/j.virusres.2014.07.012. Epub 2014 Jul 19. Virus Res. 2014. PMID: 25051147
-
Molecular biology of resistance to potato virus X in potato.Biochem Soc Symp. 1994;60:207-18. Biochem Soc Symp. 1994. PMID: 7639780 Review.
-
Potato Virus Y Emergence and Evolution from the Andes of South America to Become a Major Destructive Pathogen of Potato and Other Solanaceous Crops Worldwide.Viruses. 2020 Dec 12;12(12):1430. doi: 10.3390/v12121430. Viruses. 2020. PMID: 33322703 Free PMC article. Review.
Cited by
-
Applications of Virus-Induced Gene Silencing in Cotton.Plants (Basel). 2024 Jan 17;13(2):272. doi: 10.3390/plants13020272. Plants (Basel). 2024. PMID: 38256825 Free PMC article. Review.
-
Translating virome analyses to support biosecurity, on-farm management, and crop breeding.Front Plant Sci. 2023 Mar 14;14:1056603. doi: 10.3389/fpls.2023.1056603. eCollection 2023. Front Plant Sci. 2023. PMID: 36998684 Free PMC article. Review.
-
Limited potexvirus diversity in eastern Gulf of Mexico seagrass meadows.J Gen Virol. 2024 Jun;105(6):002004. doi: 10.1099/jgv.0.002004. J Gen Virol. 2024. PMID: 38888587 Free PMC article.
-
Role of an RNA pseudoknot involving the polyA tail in replication of Pepino mosaic potexvirus and related plant viruses.Sci Rep. 2022 Jul 7;12(1):11532. doi: 10.1038/s41598-022-15598-5. Sci Rep. 2022. PMID: 35798958 Free PMC article.
-
Prevalence and molecular characterization of important potato viruses in the Tokat province of Turkey.Mol Biol Rep. 2023 Mar;50(3):2171-2181. doi: 10.1007/s11033-022-08134-1. Epub 2022 Dec 24. Mol Biol Rep. 2023. PMID: 36565419
References
-
- Salaman R.N. The potato virus “X”: Its strains and reactions. Phil. Trans. Roy. Soc. Lond. Ser. B Biol. Sci. 1938;229:137–217.
-
- Salaman R.N. Some notes on the history of curl. Tijdschr. Plantenziekten. 1949;55:118–128. doi: 10.1007/BF01982574. - DOI
-
- Smith K.M. On the composite nature of certain potato virus diseases of the mosaic group as revealed by the use of plant indicators and selective methods of transmission. Proc. Roy. Soc. Lond. Ser. B. 1931;109:251–267.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

