Deep Learning Can Differentiate IDH-Mutant from IDH-Wild GBM
- PMID: 33918828
- PMCID: PMC8069494
- DOI: 10.3390/jpm11040290
Deep Learning Can Differentiate IDH-Mutant from IDH-Wild GBM
Abstract
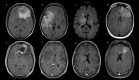
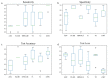
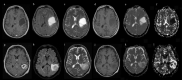
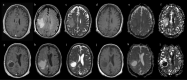
Isocitrate dehydrogenase (IDH) mutant and wildtype glioblastoma multiforme (GBM) often show overlapping features on magnetic resonance imaging (MRI), representing a diagnostic challenge. Deep learning showed promising results for IDH identification in mixed low/high grade glioma populations; however, a GBM-specific model is still lacking in the literature. Our aim was to develop a GBM-tailored deep-learning model for IDH prediction by applying convoluted neural networks (CNN) on multiparametric MRI. We selected 100 adult patients with pathologically demonstrated WHO grade IV gliomas and IDH testing. MRI sequences included: MPRAGE, T1, T2, FLAIR, rCBV and ADC. The model consisted of a 4-block 2D CNN, applied to each MRI sequence. Probability of IDH mutation was obtained from the last dense layer of a softmax activation function. Model performance was evaluated in the test cohort considering categorical cross-entropy loss (CCEL) and accuracy. Calculated performance was: rCBV (accuracy 83%, CCEL 0.64), T1 (accuracy 77%, CCEL 1.4), FLAIR (accuracy 77%, CCEL 1.98), T2 (accuracy 67%, CCEL 2.41), MPRAGE (accuracy 66%, CCEL 2.55). Lower performance was achieved on ADC maps. We present a GBM-specific deep-learning model for IDH mutation prediction, with a maximal accuracy of 83% on rCBV maps. Highest predictivity achieved on perfusion images possibly reflects the known link between IDH and neoangiogenesis through the hypoxia inducible factor.
Keywords: CBV; GBM; IDH; MRI; artificial intelligence; deep learning; high grade glioma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Tamimi A.F., Juweid M. Epidemiology and Outcome of Glioblastoma. In: De Vleeschouwer S., editor. Glioblastoma. Codon Publications; Brisbane, Australia: 2017. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

