Acquisition and Spread of Antimicrobial Resistance: A tet(X) Case Study
- PMID: 33918911
- PMCID: PMC8069840
- DOI: 10.3390/ijms22083905
Acquisition and Spread of Antimicrobial Resistance: A tet(X) Case Study
Abstract
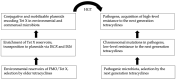
Understanding the mechanisms leading to the rise and dissemination of antimicrobial resistance (AMR) is crucially important for the preservation of power of antimicrobials and controlling infectious diseases. Measures to monitor and detect AMR, however, have been significantly delayed and introduced much later after the beginning of industrial production and consumption of antimicrobials. However, monitoring and detection of AMR is largely focused on bacterial pathogens, thus missing multiple key events which take place before the emergence and spread of AMR among the pathogens. In this regard, careful analysis of AMR development towards recently introduced antimicrobials may serve as a valuable example for the better understanding of mechanisms driving AMR evolution. Here, the example of evolution of tet(X), which confers resistance to the next-generation tetracyclines, is summarised and discussed. Initial mechanisms of resistance to these antimicrobials among pathogens were mostly via chromosomal mutations leading to the overexpression of efflux pumps. High-level resistance was achieved only after the acquisition of flavin-dependent monooxygenase-encoding genes from the environmental microbiota. These genes confer resistance to all tetracyclines, including the next-generation tetracyclines, and thus were termed tet(X). ISCR2 and IS26, as well as a variety of conjugative and mobilizable plasmids of different incompatibility groups, played an essential role in the acquisition of tet(X) genes from natural reservoirs and in further dissemination among bacterial commensals and pathogens. This process, which took place within the last decade, demonstrates how rapidly AMR evolution may progress, taking away some drugs of last resort from our arsenal.
Keywords: antimicrobial resistance; horizontal gene transfer; mobile genetic elements; natural reservoirs; next-generation tetracyclines; tetracyclines.
Conflict of interest statement
The author declares no conflict of interest.
Figures


Similar articles
-
Unravelling the menace: detection of antimicrobial resistance in aquaculture.Lett Appl Microbiol. 2020 Jul;71(1):26-38. doi: 10.1111/lam.13292. Epub 2020 Apr 22. Lett Appl Microbiol. 2020. PMID: 32248555 Review.
-
Horizontal Dissemination of Antimicrobial Resistance Determinants in Multiple Salmonella Serotypes following Isolation from the Commercial Swine Operation Environment after Manure Application.Appl Environ Microbiol. 2017 Sep 29;83(20):e01503-17. doi: 10.1128/AEM.01503-17. Print 2017 Oct 15. Appl Environ Microbiol. 2017. PMID: 28802274 Free PMC article.
-
Molecular Insights into Antimicrobial Resistance Traits of Commensal Human Gut Microbiota.Microb Ecol. 2019 Feb;77(2):546-557. doi: 10.1007/s00248-018-1228-7. Epub 2018 Jul 16. Microb Ecol. 2019. PMID: 30009332
-
Evolution of horizontal transmission in antimicrobial resistance plasmids.Microbiology (Reading). 2022 Jul;168(7). doi: 10.1099/mic.0.001214. Microbiology (Reading). 2022. PMID: 35849537 Review.
-
Genetics of antimicrobial resistance.Anim Biotechnol. 2006;17(2):111-24. doi: 10.1080/10495390600957092. Anim Biotechnol. 2006. PMID: 17127523 Review.
Cited by
-
The ISVsa3-ORF2-abh-tet(X4) circular intermediate-mediated transmission of tigecycline resistance in Escherichia coli isolates from duck farms.Front Cell Infect Microbiol. 2024 Aug 30;14:1444031. doi: 10.3389/fcimb.2024.1444031. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39282498 Free PMC article.
-
The study of honokiol as a natural product-based antimicrobial agent and its potential interaction with FtsZ protein.Front Microbiol. 2024 Jul 22;15:1361508. doi: 10.3389/fmicb.2024.1361508. eCollection 2024. Front Microbiol. 2024. PMID: 39104591 Free PMC article.
-
Aptamer-based diagnostic and therapeutic approaches in animals: Current potential and challenges.Saudi J Biol Sci. 2021 Sep;28(9):5081-5093. doi: 10.1016/j.sjbs.2021.05.031. Epub 2021 May 20. Saudi J Biol Sci. 2021. PMID: 34466086 Free PMC article. Review.
-
Low-Level Tetracycline Resistance Gene tet(O)_3 in Campylobacter jejuni.Antibiotics (Basel). 2023 Feb 21;12(3):426. doi: 10.3390/antibiotics12030426. Antibiotics (Basel). 2023. PMID: 36978293 Free PMC article.
-
Mobile Tigecycline Resistance: An Emerging Health Catastrophe Requiring Urgent One Health Global Intervention.Front Microbiol. 2022 Aug 1;13:808744. doi: 10.3389/fmicb.2022.808744. eCollection 2022. Front Microbiol. 2022. PMID: 35979498 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

