Broad-Spectrum Antiviral Activity of 3D8, a Nucleic Acid-Hydrolyzing Single-Chain Variable Fragment (scFv), Targeting SARS-CoV-2 and Multiple Coronaviruses In Vitro
- PMID: 33918914
- PMCID: PMC8068894
- DOI: 10.3390/v13040650
Broad-Spectrum Antiviral Activity of 3D8, a Nucleic Acid-Hydrolyzing Single-Chain Variable Fragment (scFv), Targeting SARS-CoV-2 and Multiple Coronaviruses In Vitro
Abstract
The virus behind the current pandemic, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the etiology of novel coronavirus disease (COVID-19) and poses a critical public health threat worldwide. Effective therapeutics and vaccines against multiple coronaviruses remain unavailable. Single-chain variable fragment (scFv), a recombinant antibody, exhibits broad-spectrum antiviral activity against DNA and RNA viruses owing to its nucleic acid-hydrolyzing property. The antiviral activity of 3D8 scFv against SARS-CoV-2 and other coronaviruses was evaluated in Vero E6 cell cultures. Viral growth was quantified with quantitative RT-qPCR and plaque assay. The nucleic acid-hydrolyzing activity of 3D8 was assessed through abzyme assays of in vitro viral transcripts and cell viability was determined by MTT assay. We found that 3D8 inhibited the replication of SARS-CoV-2, human coronavirus OC43 (HCoV-OC43), and porcine epidemic diarrhea virus (PEDV). Our results revealed the prophylactic and therapeutic effects of 3D8 scFv against SARS-CoV-2 in Vero E6 cells. Immunoblot and plaque assays showed the reduction of coronavirus nucleoproteins and infectious particles, respectively, in 3D8 scFv-treated cells. These data demonstrate the broad-spectrum antiviral activity of 3D8 against SARS-CoV-2 and other coronaviruses. Thus, it could be considered a potential antiviral countermeasure against SARS-CoV-2 and zoonotic coronaviruses.
Keywords: 3D8 scFv; COVID-19; SARS-CoV-2; coronaviruses; single-chain variable fragment.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

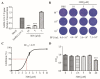

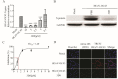
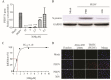

Similar articles
-
An RNA-hydrolyzing recombinant minibody prevents both influenza A virus and coronavirus in co-infection models.Sci Rep. 2024 Apr 11;14(1):8472. doi: 10.1038/s41598-024-52810-0. Sci Rep. 2024. PMID: 38605110 Free PMC article.
-
Antiviral Activity of Umifenovir In Vitro against a Broad Spectrum of Coronaviruses, Including the Novel SARS-CoV-2 Virus.Viruses. 2021 Aug 23;13(8):1665. doi: 10.3390/v13081665. Viruses. 2021. PMID: 34452529 Free PMC article.
-
An RNA-hydrolyzing recombinant antibody exhibits an antiviral activity against classical swine fever virus.Biochem Biophys Res Commun. 2010 May 14;395(4):484-9. doi: 10.1016/j.bbrc.2010.04.032. Epub 2010 Apr 9. Biochem Biophys Res Commun. 2010. PMID: 20382124
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
-
Valinomycin as a potential antiviral agent against coronaviruses: A review.Biomed J. 2020 Oct;43(5):414-423. doi: 10.1016/j.bj.2020.08.006. Epub 2020 Aug 11. Biomed J. 2020. PMID: 33012699 Free PMC article. Review.
Cited by
-
An RNA-hydrolyzing recombinant minibody prevents both influenza A virus and coronavirus in co-infection models.Sci Rep. 2024 Apr 11;14(1):8472. doi: 10.1038/s41598-024-52810-0. Sci Rep. 2024. PMID: 38605110 Free PMC article.
-
A Novel Approach of Antiviral Drugs Targeting Viral Genomes.Microorganisms. 2022 Jul 31;10(8):1552. doi: 10.3390/microorganisms10081552. Microorganisms. 2022. PMID: 36013970 Free PMC article. Review.
-
Progress of Research into Novel Drugs and Potential Drug Targets against Porcine Pseudorabies Virus.Viruses. 2022 Aug 11;14(8):1753. doi: 10.3390/v14081753. Viruses. 2022. PMID: 36016377 Free PMC article. Review.
-
Direct conversion of a general antibody to its catalytic antibody and corresponding applications -Importance and role of Pro95 in CDR-3.Proc Jpn Acad Ser B Phys Biol Sci. 2023;99(6):155-172. doi: 10.2183/pjab.99.010. Proc Jpn Acad Ser B Phys Biol Sci. 2023. PMID: 37331814 Free PMC article. Review.
-
A Therapeutically Active Minibody Exhibits an Antiviral Activity in Oseltamivir-Resistant Influenza-Infected Mice via Direct Hydrolysis of Viral RNAs.Viruses. 2022 May 21;14(5):1105. doi: 10.3390/v14051105. Viruses. 2022. PMID: 35632846 Free PMC article.
References
-
- Timothy P., Sheahan A.C.S., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., 3rd, Stevens L.J., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12:eabb5883. - PMC - PubMed
-
- Pruijssers A.J., George A.S., Schafer A., Leist S.R., Gralinksi L.E., Dinnon K.H., 3rd, Yount B.L., Agostini M.L., Stevens L.J., Chappell J.D., et al. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020;32:107940–107950. doi: 10.1016/j.celrep.2020.107940. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

