Sequence, Chromatin and Evolution of Satellite DNA
- PMID: 33919233
- PMCID: PMC8122249
- DOI: 10.3390/ijms22094309
Sequence, Chromatin and Evolution of Satellite DNA
Abstract
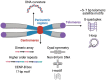
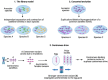
Satellite DNA consists of abundant tandem repeats that play important roles in cellular processes, including chromosome segregation, genome organization and chromosome end protection. Most satellite DNA repeat units are either of nucleosomal length or 5-10 bp long and occupy centromeric, pericentromeric or telomeric regions. Due to high repetitiveness, satellite DNA sequences have largely been absent from genome assemblies. Although few conserved satellite-specific sequence motifs have been identified, DNA curvature, dyad symmetries and inverted repeats are features of various satellite DNAs in several organisms. Satellite DNA sequences are either embedded in highly compact gene-poor heterochromatin or specialized chromatin that is distinct from euchromatin. Nevertheless, some satellite DNAs are transcribed into non-coding RNAs that may play important roles in satellite DNA function. Intriguingly, satellite DNAs are among the most rapidly evolving genomic elements, such that a large fraction is species-specific in most organisms. Here we describe the different classes of satellite DNA sequences, their satellite-specific chromatin features, and how these features may contribute to satellite DNA biology and evolution. We also discuss how the evolution of functional satellite DNA classes may contribute to speciation in plants and animals.
Keywords: CENP-A; H3K9me3; centromeres; heterochromatin; non-B-form DNA; repetitive DNA; telomeres.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

