Transcriptome Profiling of Cucumber (Cucumis sativus L.) Early Response to Pseudomonas syringae pv. lachrymans
- PMID: 33919557
- PMCID: PMC8072787
- DOI: 10.3390/ijms22084192
Transcriptome Profiling of Cucumber (Cucumis sativus L.) Early Response to Pseudomonas syringae pv. lachrymans
Abstract
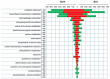
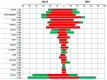
Bacterial angular leaf spot disease (ALS) caused by Pseudomonas syringae pv. lachrymans (Psl) is one of the biological factors limiting cucumber open-field production. The goal of this study was to characterize cytological and transcriptomic response of cucumber to this pathogen. Plants of two inbred lines, B10 (susceptible) and Gy14 (resistant), were grown, and leaves were inoculated with highly virulent Psl strain 814/98 under growth chamber conditions. Microscopic and transcriptional evaluations were performed at three time points: before, 1 and 3 days post inoculation (dpi). Investigated lines showed distinct response to Psl. At 1 dpi bacterial colonies were surrounded by necrotized mesophyll cells. At 3 dpi, in the susceptible B10 line bacteria were in contact with degraded cells, whereas cells next to bacteria in the resistant Gy14 line were plasmolyzed, but apparently still alive and functional. Additionally, the level of H2O2 production was higher in resistant Gy14 plants than in B10 at both examined time points. In RNA sequencing more than 18,800 transcripts were detected in each sample. As many as 1648 and 2755 differentially expressed genes (DEGs) at 1 dpi as well as 2992 and 3141 DEGs at 3 dpi were identified in B10 and Gy14, respectively. DEGs were characterized in terms of functional categories. Resistant line Gy14 showed massive transcriptomic response to Psl at 1 dpi compared to susceptible line B10, while a similar number of DEGs was detected for both lines at 3 dpi. This suggests that dynamic transcriptomic response to the invading pathogen may be related with host resistance. This manuscript provides the first transcriptomic data on cucumber infected with the pathovar lachrymans and helps to elucidate resistance mechanism against ALS disease.
Keywords: DEGs; angular leaf spot disease; defense response; transcription factors.
Conflict of interest statement
The authors state no conflict of interest.
Figures





Similar articles
-
Exogenous melatonin improves the resistance to cucumber bacterial angular leaf spot caused by Pseudomonas syringae pv. Lachrymans.Physiol Plant. 2022 May;174(3):e13724. doi: 10.1111/ppl.13724. Physiol Plant. 2022. PMID: 35611707
-
CsMYB60 confers cucumber resistance to Pseudomonas syringae pv. lachrymans through both receptor-like kinase CsFLS2-dependent and -independent pathways.Plant J. 2025 Jun;122(5):e70283. doi: 10.1111/tpj.70283. Plant J. 2025. PMID: 40513016
-
Genetic background of host-pathogen interaction between Cucumis sativus L. and Pseudomonas syringae pv. lachrymans.J Appl Genet. 2009;50(1):1-7. doi: 10.1007/BF03195645. J Appl Genet. 2009. PMID: 19193976 Review.
-
Genetic mapping of psl locus and quantitative trait loci for angular leaf spot resistance in cucumber (Cucumis sativus L.).Mol Breed. 2018;38(9):111. doi: 10.1007/s11032-018-0866-2. Epub 2018 Aug 17. Mol Breed. 2018. PMID: 30174539 Free PMC article.
-
Salicylic acid and cysteine contribute to arbutin-induced alleviation of angular leaf spot disease development in cucumber.J Plant Physiol. 2015 Jun 1;181:9-13. doi: 10.1016/j.jplph.2015.03.017. Epub 2015 Apr 17. J Plant Physiol. 2015. PMID: 25955697
Cited by
-
Genome-Wide Analysis of Exocyst Complex Subunit Exo70 Gene Family in Cucumber.Int J Mol Sci. 2023 Jun 30;24(13):10929. doi: 10.3390/ijms241310929. Int J Mol Sci. 2023. PMID: 37446106 Free PMC article.
-
Brassinosteroids in Cucurbits: Modulators of Plant Growth Architecture and Stress Response.Int J Mol Sci. 2025 Jul 26;26(15):7234. doi: 10.3390/ijms26157234. Int J Mol Sci. 2025. PMID: 40806367 Free PMC article. Review.
-
Serial-Omics and Molecular Function Study Provide Novel Insight into Cucumber Variety Improvement.Plants (Basel). 2022 Jun 20;11(12):1609. doi: 10.3390/plants11121609. Plants (Basel). 2022. PMID: 35736760 Free PMC article. Review.
-
Pseudomonas syringae Pathovar syringae Infection Reveals Different Defense Mechanisms in Two Sweet Cherry Cultivars.Plants (Basel). 2024 Dec 31;14(1):87. doi: 10.3390/plants14010087. Plants (Basel). 2024. PMID: 39795347 Free PMC article.
-
Transcriptome profiling of Toona ciliata young stems in response to Hypsipyla robusta Moore.Front Plant Sci. 2022 Aug 25;13:950945. doi: 10.3389/fpls.2022.950945. eCollection 2022. Front Plant Sci. 2022. PMID: 36105698 Free PMC article.
References
-
- FAOSTAT Database. [(accessed on 17 January 2021)]; Available online: http://www.fao.org/faostat/en/#home.
-
- Ainsworth G.C. Introduction to the History of Plant Pathology. Cambridge University Press; Cambridge, UK: 1981. p. 68.
-
- Janse J.D. Phytobacteriology: Principles and Practice. CABI Publishing; Wallingford, UK: 2005. pp. 252–289.
-
- Bradbury J.F. Guide to Plant Pathogenic Bacteria. CAB International Mycological Institute; Kew, Surrey, UK: 1986.
-
- Bhat N.A., Bhat K.A., Zargar M.Y., Teli M.A., Nazir M., Zargar S.M. Current status of angular leaf spot (Pseudomonas syringae pv. lachrymans) of cucumber: A review. Int. J. Curr. Res. 2010;8:1–11.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

