Characterization of a Novel Group of Listeria Phages That Target Serotype 4b Listeria monocytogenes
- PMID: 33919793
- PMCID: PMC8070769
- DOI: 10.3390/v13040671
Characterization of a Novel Group of Listeria Phages That Target Serotype 4b Listeria monocytogenes
Abstract
Listeria monocytogenes serotype 4b strains are the most prevalent clinical isolates and are widely found in food processing environments. Bacteriophages are natural viral predators of bacteria and are a promising biocontrol agent for L. monocytogenes. The aims of this study were to characterize phages that specifically infect serotype 4b strains and to assess their ability to inhibit the growth of serotype 4b strains. Out of 120 wild Listeria phages, nine phages were selected based on their strong lytic activity against the model serotype 4b strain F2365. These nine phages can be divided into two groups based on their morphological characteristics and host range. Comparison to previously characterized phage genomes revealed one of these groups qualifies to be defined as a novel species. Phages LP-020, LP-027, and LP-094 were selected as representatives of these two groups of phages for further characterization through one-step growth curve and inhibition of serotype 4b L. monocytogenes experiments. Listeria phages that target serotype 4b showed an inhibitory effect on the growth of F2365 and other serotype 4 strains and may be useful for biocontrol of L.monocytogenes in food processing environments.
Keywords: Listeria monocytogenes; bacteriophage; phage; serotype 4b.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
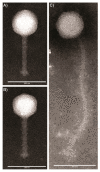

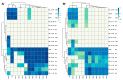
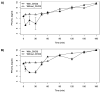

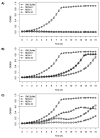
References
-
- Karthikeyan R., Gayathri P., Gunasekaran P., Jagannadham M.V., Rajendhran J. Comprehensive proteomic analysis and pathogenic role of membrane vesicles of Listeria monocytogenes serotype 4b reveals proteins associated with virulence and their possible interaction with host. Int. J. Med. Microbiol. 2019;309:199–212. doi: 10.1016/j.ijmm.2019.03.008. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

