Genome-Wide Identification and Analysis of Nilaparvata lugens microRNAs during Challenge with the Entomopathogenic Fungus Metarhizium anisopliae
- PMID: 33919937
- PMCID: PMC8070897
- DOI: 10.3390/jof7040295
Genome-Wide Identification and Analysis of Nilaparvata lugens microRNAs during Challenge with the Entomopathogenic Fungus Metarhizium anisopliae
Abstract
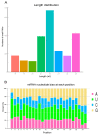
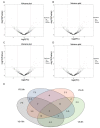
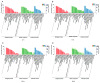
The resistance of the notorious rice pest Nilaparvata lugens to many insecticides has caused significant concerns. Our previous study demonstrated that the fungus Metarhizium anisopliae CQMa421 shows great potential for the control of this pest, but the interactions between them are still unclear. Thus, we further investigated fungal infection-related microRNAs (miRNAs) in N. lugens during M. anisopliae CQMa421 challenge using Illumina sequencing. In this study, we constructed twenty-four small RNA libraries over different time courses (i.e., 4 h, 8 h, 16 h, and 24 h). A total of 478.62 M clean reads were collected, with each sample producing more than 13.37 M reads, after the removal of low-quality reads. We identified 2324 miRNAs and their 11,076 target genes within the twenty-four libraries by bioinformatics analysis. Differentially expressed miRNAs (DEmiRNAs), including 58 (32 upregulated vs. 26 downregulated), 62 (30 upregulated vs. 32 downregulated), 126 (71 upregulated vs. 55 downregulated), and 109 (40 upregulated vs. 69 downregulated) DEmiRNAs were identified at 4 h, 8 h, 16 h, and 24 h post-infection, respectively. We further conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis to predict the functions of all target genes of DEmiRNAs. These DEmiRNAs targets identified during 24 h of infection were primarily involved in energy metabolism, lysine degradation, the FoxO signaling pathway, ubiquitin-mediated proteolysis, the mRNA surveillance pathway, and the MAPK signaling pathway. Taken together, our results provide essential information for further study of the interactions between the entomopathogenic fungus M. anisopliae and N. lugens at the posttranscriptional level.
Keywords: Metarhizium anisopliae; Nilaparvata lugens; entomopathogenic fungus; fungal infection; microRNAs; pest control.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The Entomopathogenic Fungus Metarhizium anisopliae Affects Feeding Preference of Sogatella furcifera and Its Potential Targets' Identification.J Fungi (Basel). 2022 May 15;8(5):506. doi: 10.3390/jof8050506. J Fungi (Basel). 2022. PMID: 35628761 Free PMC article.
-
Transcriptomic Analysis of the Brown Planthopper, Nilaparvata lugens, at Different Stages after Metarhizium anisopliae Challenge.Insects. 2020 Feb 24;11(2):139. doi: 10.3390/insects11020139. Insects. 2020. PMID: 32102435 Free PMC article.
-
Effects of the Entomopathogenic Fungus Metarhizium anisopliae on the Mortality and Immune Response of Locusta migratoria.Insects. 2019 Dec 31;11(1):36. doi: 10.3390/insects11010036. Insects. 2019. PMID: 31906210 Free PMC article.
-
Characterization and comparative profiling of the small RNA transcriptomes in the Hemipteran insect Nilaparvata lugens.Gene. 2016 Dec 20;595(1):83-91. doi: 10.1016/j.gene.2016.09.042. Epub 2016 Sep 28. Gene. 2016. PMID: 27693372
-
Severe fungal sclerokeratitis caused by Metarhizium anisopliae: a case report and literature review.Mycoses. 2015 Feb;58(2):88-92. doi: 10.1111/myc.12279. Epub 2015 Jan 15. Mycoses. 2015. PMID: 25590990 Review.
Cited by
-
Versatile Roles of Microbes and Small RNAs in Rice and Planthopper Interactions.Plant Pathol J. 2022 Oct;38(5):432-448. doi: 10.5423/PPJ.RW.07.2022.0090. Epub 2022 Oct 1. Plant Pathol J. 2022. PMID: 36221916 Free PMC article. Review.
-
The Entomopathogenic Fungus Metarhizium anisopliae Affects Feeding Preference of Sogatella furcifera and Its Potential Targets' Identification.J Fungi (Basel). 2022 May 15;8(5):506. doi: 10.3390/jof8050506. J Fungi (Basel). 2022. PMID: 35628761 Free PMC article.
References
-
- Lou Y.G., Zhang G.R., Zhang W.Q., Hu Y., Zhang J. Biological control of rice insect pests in China. Biol. Control. 2013;67:8–20. doi: 10.1016/j.biocontrol.2013.06.011. - DOI
-
- Cheng J. Rice planthopper problems and relevant causes in China. Rice. 2009;1:1–32.
-
- Wang Y., Tang M., Hao P., Yang Z., Zhu L., He G. Penetration into rice tissues by brown planthopper and fine structure of the salivary sheaths. Entomol. Exp. Appl. 2010;129:295–307. doi: 10.1111/j.1570-7458.2008.00785.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

