Fish-Ing for Enhancers in the Heart
- PMID: 33920121
- PMCID: PMC8069060
- DOI: 10.3390/ijms22083914
Fish-Ing for Enhancers in the Heart
Abstract
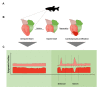
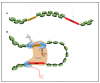
Precise control of gene expression is crucial to ensure proper development and biological functioning of an organism. Enhancers are non-coding DNA elements which play an essential role in regulating gene expression. They contain specific sequence motifs serving as binding sites for transcription factors which interact with the basal transcription machinery at their target genes. Heart development is regulated by intricate gene regulatory network ensuring precise spatiotemporal gene expression program. Mutations affecting enhancers have been shown to result in devastating forms of congenital heart defect. Therefore, identifying enhancers implicated in heart biology and understanding their mechanism is key to improve diagnosis and therapeutic options. Despite their crucial role, enhancers are poorly studied, mainly due to a lack of reliable way to identify them and determine their function. Nevertheless, recent technological advances have allowed rapid progress in enhancer discovery. Model organisms such as the zebrafish have contributed significant insights into the genetics of heart development through enabling functional analyses of genes and their regulatory elements in vivo. Here, we summarize the current state of knowledge on heart enhancers gained through studies in model organisms, discuss various approaches to discover and study their function, and finally suggest methods that could further advance research in this field.
Keywords: enhancers; heart development; heart regeneration; model organism; transcription factors; zebrafish.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
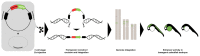
Similar articles
-
Evaluating the transcriptional regulators of arterial gene expression via a catalogue of characterized arterial enhancers.Elife. 2025 Jan 17;14:e102440. doi: 10.7554/eLife.102440. Elife. 2025. PMID: 39819837 Free PMC article.
-
Conserved and non-conserved enhancers direct tissue specific transcription in ancient germ layer specific developmental control genes.BMC Dev Biol. 2011 Oct 20;11:63. doi: 10.1186/1471-213X-11-63. BMC Dev Biol. 2011. PMID: 22011226 Free PMC article.
-
Identification of activated enhancers and linked transcription factors in breast, prostate, and kidney tumors by tracing enhancer networks using epigenetic traits.Epigenetics Chromatin. 2016 Nov 9;9:50. doi: 10.1186/s13072-016-0102-4. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27833659 Free PMC article.
-
Towards genome-wide prediction and characterization of enhancers in plants.Biochim Biophys Acta Gene Regul Mech. 2017 Jan;1860(1):131-139. doi: 10.1016/j.bbagrm.2016.06.006. Epub 2016 Jun 16. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 27321818 Review.
-
Distal enhancers: new insights into heart development and disease.Trends Cell Biol. 2014 May;24(5):294-302. doi: 10.1016/j.tcb.2013.10.008. Epub 2013 Dec 7. Trends Cell Biol. 2014. PMID: 24321408 Review.
Cited by
-
Identification of ancestral gnathostome Gli3 enhancers with activity in mammals.Dev Growth Differ. 2024 Jan;66(1):75-88. doi: 10.1111/dgd.12901. Epub 2023 Dec 20. Dev Growth Differ. 2024. PMID: 37925606 Free PMC article.
-
Identification and Copy Number Variant Analysis of Enhancer Regions of Genes Causing Spinocerebellar Ataxia.Int J Mol Sci. 2024 Oct 18;25(20):11205. doi: 10.3390/ijms252011205. Int J Mol Sci. 2024. PMID: 39456985 Free PMC article.
-
The combinatorial binding syntax of transcription factors in forebrain-specific enhancers.Biol Open. 2025 Feb 15;14(2):BIO061751. doi: 10.1242/bio.061751. Epub 2025 Feb 19. Biol Open. 2025. PMID: 39976127 Free PMC article.
-
Advances in Cardiac Development and Regeneration Using Zebrafish as a Model System for High-Throughput Research.J Dev Biol. 2021 Sep 25;9(4):40. doi: 10.3390/jdb9040040. J Dev Biol. 2021. PMID: 34698193 Free PMC article. Review.
-
Exposure to tris (1,3-dichloro-2-propyl) phosphate affects the embryonic cardiac development of Oryzias melastigma.Heliyon. 2024 Feb 1;10(3):e25554. doi: 10.1016/j.heliyon.2024.e25554. eCollection 2024 Feb 15. Heliyon. 2024. PMID: 38327441 Free PMC article.
References
-
- Ounzain S., Pezzuto I., Micheletti R., Burdet F., Sheta R., Nemir M., Gonzales C., Sarre A., Alexanian M., Blow M.J., et al. Functional Importance of Cardiac Enhancer-Associated Noncoding RNAs in Heart Development and Disease. J. Mol. Cell. Cardiol. 2014;76:55–70. doi: 10.1016/j.yjmcc.2014.08.009. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

