Agrobacterium-Mediated Capsicum annuum Gene Editing in Two Cultivars, Hot Pepper CM334 and Bell Pepper Dempsey
- PMID: 33920210
- PMCID: PMC8070316
- DOI: 10.3390/ijms22083921
Agrobacterium-Mediated Capsicum annuum Gene Editing in Two Cultivars, Hot Pepper CM334 and Bell Pepper Dempsey
Abstract
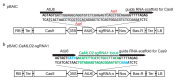
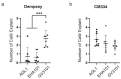
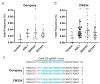
Peppers (Capsicum annuum L.) are the most widespread and cultivated species of Solanaceae in subtropical and temperate countries. These vegetables are economically attractive worldwide. Although whole-genome sequences of peppers and genome-editing tools are currently available, the precision editing of peppers is still in its infancy because of the lack of a stable pepper transformation method. Here, we employed three Agrobacterium tumefaciens strains-AGL1, EHA101, and GV3101-to investigate which Agrobacterium strain could be used for pepper transformation. Hot pepper CM334 and bell pepper Dempsey were chosen in this study. Agrobacterium tumefaciens GV3101 induced the highest number of calli in cv. Dempsey. All three strains generated similar numbers of calli for cv. CM334. We optimized a suitable concentration of phosphinothricin (PPT) to select a CRISPR/Cas9 binary vector (pBAtC) for both pepper types. Finally, we screened transformed calli for PPT resistance (1 and 5 mg/L PPT for cv. CM334 and Dempsey, respectively). These selected calli showed different indel frequencies from the non-transformed calli. However, the primary indel pattern was consistent with a 1-bp deletion at the target locus of the C. annuumMLO gene (CaMLO2). These results demonstrate the different sensitivity between cv. CM334 and Dempsey to A. tumefaciens-mediated callus induction, and a differential selection pressure of PPT via pBAtC binary vector.
Keywords: Agrobacterium tumefaciens; CRISPR/Cas9; CaMLO2; Capsicum annuum CM334; Capsicum annuum Dempsey; pBAtC binary vector.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Lyngkjær M.F., Newton A.C., Atzema J.L., Baker S.J. The Barley mlo-gene: An important powdery mildew resistance source. Agronomie. 2000;20:745–756. doi: 10.1051/agro:2000173. - DOI
-
- Acevedo-Garcia J., Gruner K., Reinstädler A., Kemen A., Kemen E., Cao L., Takken F.L.W., Reitz M.U., Schäfer P., O’Connell R.J., et al. The powdery mildew-resistant Arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Sci. Rep. 2017;7:9319. doi: 10.1038/s41598-017-07188-7. - DOI - PMC - PubMed
MeSH terms
Grants and funding
- Project No. PJ01477602/the New Breeding Technologies Development Program, Rural Devel-opment Administration (RDA)
- NRF-2018R1A2B6006233/the Basic Science Research Program of the National Research Foundation of Korea
- 2019 URP/the Korea Foundation for the Advancement of Science & Creativity (KOFAC), and funded by the Korean Government (MOE)
LinkOut - more resources
Full Text Sources
Other Literature Sources

