Outer Membrane Vesicle Production by Helicobacter pylori Represents an Approach for the Delivery of Virulence Factors CagA, VacA and UreA into Human Gastric Adenocarcinoma (AGS) Cells
- PMID: 33920443
- PMCID: PMC8069053
- DOI: 10.3390/ijms22083942
Outer Membrane Vesicle Production by Helicobacter pylori Represents an Approach for the Delivery of Virulence Factors CagA, VacA and UreA into Human Gastric Adenocarcinoma (AGS) Cells
Abstract
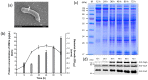
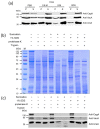
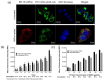
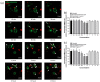
Helicobacter pylori infection is the etiology of several gastric-related diseases including gastric cancer. Cytotoxin associated gene A (CagA), vacuolating cytotoxin A (VacA) and α-subunit of urease (UreA) are three major virulence factors of H. pylori, and each of them has a distinct entry pathway and pathogenic mechanism during bacterial infection. H. pylori can shed outer membrane vesicles (OMVs). Therefore, it would be interesting to explore the production kinetics of H. pylori OMVs and its connection with the entry of key virulence factors into host cells. Here, we isolated OMVs from H. pylori 26,695 strain and characterized their properties and interaction kinetics with human gastric adenocarcinoma (AGS) cells. We found that the generation of OMVs and the presence of CagA, VacA and UreA in OMVs were a lasting event throughout different phases of bacterial growth. H. pylori OMVs entered AGS cells mainly through macropinocytosis/phagocytosis. Furthermore, CagA, VacA and UreA could enter AGS cells via OMVs and the treatment with H. pylori OMVs would cause cell death. Comparison of H. pylori 26,695 and clinical strains suggested that the production and characteristics of OMVs are not only limited to laboratory strains commonly in use, but a general phenomenon to most H. pylori strains.
Keywords: CagA; Helicobacter pylori; UreA; VacA; gastrointestinal disorders; outer membrane vesicles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Role of Helicobacter pylori virulence factors for iron acquisition from gastric epithelial cells of the host and impact on bacterial colonization.Future Microbiol. 2011 Aug;6(8):843-6. doi: 10.2217/fmb.11.75. Future Microbiol. 2011. PMID: 21861616
-
H. pylori-encoded CagA disrupts tight junctions and induces invasiveness of AGS gastric carcinoma cells via Cdx2-dependent targeting of Claudin-2.Cell Immunol. 2013 Nov-Dec;286(1-2):22-30. doi: 10.1016/j.cellimm.2013.10.008. Epub 2013 Nov 9. Cell Immunol. 2013. PMID: 24287273
-
Evaluation of apoptosis induction using PARP cleavage on gastric adenocarcinoma and fibroblast cell lines by different strains of Helicobacter pylori.Pak J Biol Sci. 2007 Nov 15;10(22):4097-102. doi: 10.3923/pjbs.2007.4097.4102. Pak J Biol Sci. 2007. PMID: 19090286
-
Influence of Helicobacter pylori virulence factors CagA and VacA on pathogenesis of gastrointestinal disorders.Microb Pathog. 2018 Apr;117:43-48. doi: 10.1016/j.micpath.2018.02.016. Epub 2018 Feb 9. Microb Pathog. 2018. PMID: 29432909 Review.
-
Clinical relevance of Helicobacter pylori vacA and cagA genotypes in gastric carcinoma.Best Pract Res Clin Gastroenterol. 2014 Dec;28(6):1003-15. doi: 10.1016/j.bpg.2014.09.004. Epub 2014 Oct 2. Best Pract Res Clin Gastroenterol. 2014. PMID: 25439067 Review.
Cited by
-
Both extracellular vesicles from helicobacter pylori-infected cells and helicobacter pylori outer membrane vesicles are involved in gastric/extragastric diseases.Eur J Med Res. 2023 Nov 6;28(1):484. doi: 10.1186/s40001-023-01458-z. Eur J Med Res. 2023. PMID: 37932800 Free PMC article. Review.
-
Bacterial Membrane Vesicles: Physiological Roles, Infection Immunology, and Applications.Adv Sci (Weinh). 2023 Sep;10(25):e2301357. doi: 10.1002/advs.202301357. Epub 2023 Jun 25. Adv Sci (Weinh). 2023. PMID: 37357142 Free PMC article. Review.
-
Helicobacter pylori Outer Membrane Vesicles: Biogenesis, Composition, and Biological Functions.Int J Biol Sci. 2024 Jul 15;20(10):4029-4043. doi: 10.7150/ijbs.94156. eCollection 2024. Int J Biol Sci. 2024. PMID: 39113715 Free PMC article. Review.
-
Selective Inhibition of Helicobacter pylori Carbonic Anhydrases by Carvacrol and Thymol Could Impair Biofilm Production and the Release of Outer Membrane Vesicles.Int J Mol Sci. 2021 Oct 27;22(21):11583. doi: 10.3390/ijms222111583. Int J Mol Sci. 2021. PMID: 34769015 Free PMC article.
-
A new isolation method for bacterial extracellular vesicles providing greater purity and improved proteomic detection of vesicle proteins.J Extracell Biol. 2023 Apr 25;2(5):e84. doi: 10.1002/jex2.84. eCollection 2023 May. J Extracell Biol. 2023. PMID: 38938280 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

