Escherichia coli Affects Expression of Circadian Clock Genes in Human Hepatoma Cells
- PMID: 33920679
- PMCID: PMC8073551
- DOI: 10.3390/microorganisms9040869
Escherichia coli Affects Expression of Circadian Clock Genes in Human Hepatoma Cells
Abstract
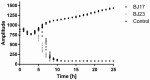
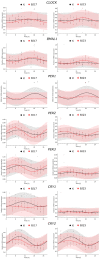
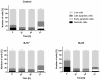
Recent research has indicated that dysbiosis of the gut microbiota can lead to an altered circadian clock of the mammalian host. Herein we developed an original system that allows real-time circadian studies of human HepG2 hepatoma cells co-cultured with bacteria. The HepG2 cells with stably integrated firefly luciferase reporter under the control of PERIOD2 promoter were co-cultured with E. coli strains isolated from human fecal samples from healthy individuals. The two E. coli strains differ in the phylogenetic group and the number of ExPEC virulence-associated genes: BJ17 has only two, and BJ23 has 15 of 23 tested. In the first 24 h, the E. coli BJ17 affected the HepG2 circadian clock more than BJ23. Cosinor analysis shows a statistically significant change in the amplitude of PER1 and 2 and the phase advance of PER3. A high percentage of necrotic and apoptotic cells occurred at 72 h, while a correlation between the number of ExPEC genes and the influence on the HepG2 core clock gene expression was observed. Our study reveals that the E. coli genetic background is important for the effect on the mammalian circadian clock genes, indicating possible future use of probiotic E. coli strains to influence the host circadian clock.
Keywords: Escherichia coli; HepG2; circadian clock; circadian rhythm; intestinal microbiota.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Leone V., Gibbons S.M., Martinez K., Hutchison A.L., Huang E.Y., Cham C.M., Pierre J.F., Heneghan A.F., Nadimpalli A., Hubert N., et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

