A Label-Free DNA-Immunosensor Based on Aminated rGO Electrode for the Quantification of DNA Methylation
- PMID: 33921234
- PMCID: PMC8070590
- DOI: 10.3390/nano11040985
A Label-Free DNA-Immunosensor Based on Aminated rGO Electrode for the Quantification of DNA Methylation
Abstract
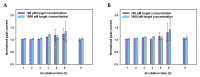
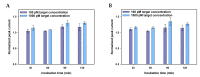
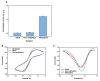
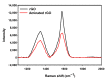
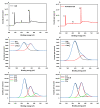
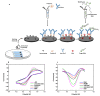
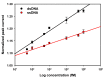
In this work, we developed a sandwich DNA-immunosensor for quantification of the methylated tumour suppressor gene O-6-methylguanine-DNA methyltransferase (MGMT), which is a potential biomarker for brain tumours and breast cancer. The biosensor is based on aminated reduced graphene oxide electrode, which is achieved by ammonium hydroxide chemisorption and anti-5-methylcytosine (anti-5mC) as a methylation bioreceptor. The target single-strand (ss) MGMT oligonucleotide is first recognised by its hybridisation with complementary DNA to form double-stranded (ds) MGMT, which is then captured by anti-5mC on the electrode surface due to the presence of methylation. Raman spectroscopy, X-ray photoelectron spectroscopy (XPS) and Scanning electron microscopy (SEM) techniques were used to characterise the electrode surface. Cyclic voltammetry (CV) and differential pulse voltammetry (DPV) techniques were used for electrochemical measurements. Under optimised conditions, the proposed biosensor is able to quantify a linear range of concentrations of the MGMT gene from 50 fM to 100 pM with a limit of detection (LOD) of 12 fM. The sandwich design facilitates the simultaneous recognition and quantification of DNA methylation, and the amination significantly improves the sensitivity of the biosensor. This biosensor is label-, bisulfite- and PCR-free and has a simple design for cost-efficient production. It can also be tailor-made to detect other methylated genes, which makes it a promising detection platform for DNA methylation-related disease diagnosis and prognosis.
Keywords: MGMT gene; NH2 chemisorption; amination; quantification of DNA methylation; reduced graphene oxide (rGO).
Conflict of interest statement
The authors declare no conflict of interest.
Figures




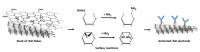
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

