Nonlinear T-Wave Time Warping-Based Sensing Model for Non-Invasive Personalised Blood Potassium Monitoring in Hemodialysis Patients: A Pilot Study
- PMID: 33921468
- PMCID: PMC8069025
- DOI: 10.3390/s21082710
Nonlinear T-Wave Time Warping-Based Sensing Model for Non-Invasive Personalised Blood Potassium Monitoring in Hemodialysis Patients: A Pilot Study
Abstract
Background: End-stage renal disease patients undergoing hemodialysis (ESRD-HD) therapy are highly susceptible to malignant ventricular arrhythmias caused by undetected potassium concentration ([K+]) variations (Δ[K+]) out of normal ranges. Therefore, a reliable method for continuous, noninvasive monitoring of [K+] is crucial. The morphology of the T-wave in the electrocardiogram (ECG) reflects Δ[K+] and two time-warping-based T-wave morphological parameters, dw and its heart-rate corrected version dw,c, have been shown to reliably track Δ[K+] from the ECG. The aim of this study is to derive polynomial models relating dw and dw,c with Δ[K+], and to test their ability to reliably sense and quantify Δ[K+] values.
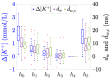
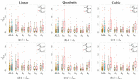
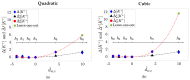
Methods: 48-hour Holter ECGs and [K+] values from six blood samples were collected from 29 ESRD-HD patients. For every patient, dw and dw,c were computed, and linear, quadratic, and cubic fitting models were derived from them. Then, Spearman's (ρ) and Pearson's (r) correlation coefficients, and the estimation error (ed) between Δ[K+] and the corresponding model-estimated values (Δ^[K+]) were calculated.
Results and discussions: Nonlinear models were the most suitable for Δ[K+] estimation, rendering higher Pearson's correlation (median 0.77 ≤r≤ 0.92) and smaller estimation error (median 0.20 ≤ed≤ 0.43) than the linear model (median 0.76 ≤r≤ 0.86 and 0.30 ≤ed≤ 0.40), even if similar Spearman's ρ were found across models (median 0.77 ≤ρ≤ 0.83).
Conclusion: Results support the use of nonlinear T-wave-based models as Δ[K+] sensors in ESRD-HD patients.
Keywords: T-wave morphology; electrocardiogram; noninvasive potassium sensing; periodic component analysis; personalised medicine; time warping.
Conflict of interest statement
The authors declare no conflict of interest. The funders had role in the design of the study but they had no role in the collection, analyses, or interpretation of data nor in the writing of the manuscript, or in the decision to publish the results.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

