Molecular Characterization of Potato Virus Y (PVY) Using High-Throughput Sequencing: Constraints on Full Genome Reconstructions Imposed by Mixed Infection Involving Recombinant PVY Strains
- PMID: 33921504
- PMCID: PMC8069754
- DOI: 10.3390/plants10040753
Molecular Characterization of Potato Virus Y (PVY) Using High-Throughput Sequencing: Constraints on Full Genome Reconstructions Imposed by Mixed Infection Involving Recombinant PVY Strains
Abstract
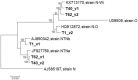
In recent years, high throughput sequencing (HTS) has brought new possibilities to the study of the diversity and complexity of plant viromes. Mixed infection of a single plant with several viruses is frequently observed in such studies. We analyzed the virome of 10 tomato and sweet pepper samples from Slovakia, all showing the presence of potato virus Y (PVY) infection. Most datasets allow the determination of the nearly complete sequence of a single-variant PVY genome, belonging to one of the PVY recombinant strains (N-Wi, NTNa, or NTNb). However, in three to-mato samples (T1, T40, and T62) the presence of N-type and O-type sequences spanning the same genome region was documented, indicative of mixed infections involving different PVY strains variants, hampering the automated assembly of PVY genomes present in the sample. The N- and O-type in silico data were further confirmed by specific RT-PCR assays targeting UTR-P1 and NIa genomic parts. Although full genomes could not be de novo assembled directly in this situation, their deep coverage by relatively long paired reads allowed their manual re-assembly using very stringent mapping parameters. These results highlight the complexity of PVY infection of some host plants and the challenges that can be met when trying to precisely identify the PVY isolates involved in mixed infection.
Keywords: PVY; Solanaceae; genome; next generation sequencing; potyvirus.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
Detection and molecular characterization of Slovak tomato isolates belonging to two recombinant strains of potato virus Y.Acta Virol. 2016;60(4):347-353. doi: 10.4149/av_2016_04_347. Acta Virol. 2016. PMID: 27928913
-
Prevalence and Strain Composition of Potato virus Y Circulating in Potato Fields in China's North-Central Province of Shanxi.Plant Dis. 2022 May;106(5):1434-1445. doi: 10.1094/PDIS-09-21-1950-RE. Epub 2022 Apr 4. Plant Dis. 2022. PMID: 34813711
-
Whole genome characterization of Potato virus Y isolates collected in the western USA and their comparison to isolates from Europe and Canada.Arch Virol. 2006 Jun;151(6):1055-74. doi: 10.1007/s00705-005-0707-6. Epub 2006 Feb 7. Arch Virol. 2006. PMID: 16463126
-
Discussion paper: The naming of Potato virus Y strains infecting potato.Arch Virol. 2008;153(1):1-13. doi: 10.1007/s00705-007-1059-1. Epub 2007 Oct 18. Arch Virol. 2008. PMID: 17943395 Review.
-
Potato Virus Y Emergence and Evolution from the Andes of South America to Become a Major Destructive Pathogen of Potato and Other Solanaceous Crops Worldwide.Viruses. 2020 Dec 12;12(12):1430. doi: 10.3390/v12121430. Viruses. 2020. PMID: 33322703 Free PMC article. Review.
Cited by
-
Molecular Footprints of Potato Virus Y Isolate Infecting Potatoes (Solanum tuberosum) in Kenya.Adv Virol. 2024 Aug 6;2024:2197725. doi: 10.1155/2024/2197725. eCollection 2024. Adv Virol. 2024. PMID: 39139708 Free PMC article.
-
High-Throughput Sequencing Discloses the Cucumber Mosaic Virus (CMV) Diversity in Slovakia and Reveals New Hosts of CMV from the Papaveraceae Family.Plants (Basel). 2022 Jun 23;11(13):1665. doi: 10.3390/plants11131665. Plants (Basel). 2022. PMID: 35807616 Free PMC article.
-
Viromes of 15 Pepper (Capsicum annuum L.) Cultivars.Int J Mol Sci. 2022 Sep 10;23(18):10507. doi: 10.3390/ijms231810507. Int J Mol Sci. 2022. PMID: 36142418 Free PMC article. Review.
-
Identification of Cacao Mild Mosaic Virus (CaMMV) and Cacao Yellow Vein-Banding Virus (CYVBV) in Cocoa (Theobroma cacao) Germplasm.Viruses. 2021 Oct 26;13(11):2152. doi: 10.3390/v13112152. Viruses. 2021. PMID: 34834959 Free PMC article.
References
-
- Lacomme C., Jacquot E. General Characteristics of Potato virus Y (PVY) and Its Impact on Potato Production, An Overview. In: Lacomme C., Glais Lv Bellstedt D., Dupuis B., Karasev A., Jacquot E., editors. Potato virus Y: Biodiversity, Pathogenicity, Epidemiology and Management. Springer; Cham, Switzerland: 2017. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

