Genomic Surveillance of Circulating SARS-CoV-2 in South East Italy: A One-Year Retrospective Genetic Study
- PMID: 33922257
- PMCID: PMC8147059
- DOI: 10.3390/v13050731
Genomic Surveillance of Circulating SARS-CoV-2 in South East Italy: A One-Year Retrospective Genetic Study
Abstract
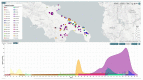
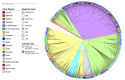
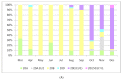
In order to provide insights into the evolutionary and epidemiological viral dynamics during the current COVID-19 pandemic in South Eastern Italy, a total of 298 genomes of SARS-CoV-2 strains collected in the Apulia and Basilicata regions, between March 2020 and January 2021, were sequenced. The genomic analysis performed on the draft genomes allowed us to assign the genetic clades and lineages of belonging to each sample and provide an overview of the main circulating viral variants. Our data showed the spread in Apulia and Basilicata of SARS-CoV-2 variants which have emerged during the second wave of infections and are being currently monitored worldwide for their increased transmission rate and their possible impact on vaccines and therapies. These results emphasize the importance of genome sequencing for the epidemiological surveillance of the new SARS-CoV-2 variants' spread.
Keywords: COVID-19; Nextstrain clade; Pangolin lineage; SARS-CoV-2; genomic surveillance; variants of concern; whole genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Mutation profile of SARS-CoV-2 genome in a sample from the first year of the pandemic in Colombia.Infect Genet Evol. 2022 Jan;97:105192. doi: 10.1016/j.meegid.2021.105192. Epub 2021 Dec 18. Infect Genet Evol. 2022. PMID: 34933126 Free PMC article.
-
Molecular Characterization and Genomic Surveillance of SARS-CoV-2 Lineages in Central India.Viruses. 2024 Oct 14;16(10):1608. doi: 10.3390/v16101608. Viruses. 2024. PMID: 39459941 Free PMC article.
-
Circulating Variants of SARS-CoV-2 Among Macedonian COVID - 19 Patients in the First Year of Pandemic.Pril (Makedon Akad Nauk Umet Odd Med Nauki). 2023 Jul 15;44(2):17-22. doi: 10.2478/prilozi-2023-0018. Print 2023 Jul 1. Pril (Makedon Akad Nauk Umet Odd Med Nauki). 2023. PMID: 37453123
-
One year into the pandemic: Short-term evolution of SARS-CoV-2 and emergence of new lineages.Infect Genet Evol. 2021 Aug;92:104869. doi: 10.1016/j.meegid.2021.104869. Epub 2021 Apr 26. Infect Genet Evol. 2021. PMID: 33915216 Free PMC article. Review.
-
SARS-CoV-2 Lineages and Sub-Lineages Circulating Worldwide: A Dynamic Overview.Chemotherapy. 2021;66(1-2):3-7. doi: 10.1159/000515340. Epub 2021 Mar 18. Chemotherapy. 2021. PMID: 33735881 Free PMC article. Review.
Cited by
-
A regional genomic surveillance program is implemented to monitor the occurrence and emergence of SARS-CoV-2 variants in Yubei District, China.Virol J. 2024 Jan 8;21(1):13. doi: 10.1186/s12985-023-02279-6. Virol J. 2024. PMID: 38191416 Free PMC article.
-
SARS-CoV-2 Pandemic Tracing in Italy Highlights Lineages with Mutational Burden in Growing Subsets.Int J Mol Sci. 2022 Apr 9;23(8):4155. doi: 10.3390/ijms23084155. Int J Mol Sci. 2022. PMID: 35456974 Free PMC article.
-
Emerging Mutations Potentially Related to SARS-CoV-2 Immune Escape: The Case of a Long-Term Patient.Life (Basel). 2021 Nov 18;11(11):1259. doi: 10.3390/life11111259. Life (Basel). 2021. PMID: 34833135 Free PMC article.
-
SARS-CoV-2 in Animal Companions: A Serosurvey in Three Regions of Southern Italy.Life (Basel). 2023 Dec 16;13(12):2354. doi: 10.3390/life13122354. Life (Basel). 2023. PMID: 38137955 Free PMC article.
-
Analysis of Genomic Characteristics of SARS-CoV-2 in Italy, 29 January to 27 March 2020.Viruses. 2022 Feb 25;14(3):472. doi: 10.3390/v14030472. Viruses. 2022. PMID: 35336879 Free PMC article.
References
-
- WHO Coronavirus Disease (COVID-19) Dashboard. [(accessed on 14 February 2021)]; Available online: https://covid19.who.int.
-
- Stefanelli P., Faggioni G., Lo Presti A., Fiore S., Marchi A., Benedetti E., Fabiani C., Anselmo A., Ciammaruconi A., Fortunato A., et al. Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: Additional clues on multiple introductions and further circulation in Europe. Euro. Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.13.2000305. - DOI - PMC - PubMed
-
- Capobianchi M.R., Rueca M., Messina F., Giombini E., Carletti F., Colavita F., Castilletti C., Lalle E., Bordi L., Vairo F., et al. Molecular characterization of SARS-CoV-2 from the first case of COVID-19 in Italy. Clin. Microbiol. Infect. 2020;26:954–956. doi: 10.1016/j.cmi.2020.03.025. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

