Characterization of a Dye-Decolorizing Peroxidase from Irpex lacteus Expressed in Escherichia coli: An Enzyme with Wide Substrate Specificity Able to Transform Lignosulfonates
- PMID: 33922393
- PMCID: PMC8145141
- DOI: 10.3390/jof7050325
Characterization of a Dye-Decolorizing Peroxidase from Irpex lacteus Expressed in Escherichia coli: An Enzyme with Wide Substrate Specificity Able to Transform Lignosulfonates
Abstract
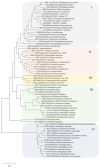
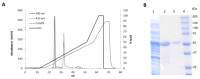
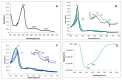
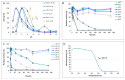
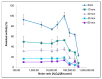
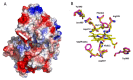
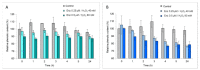
A dye-decolorizing peroxidase (DyP) from Irpex lacteus was cloned and heterologously expressed as inclusion bodies in Escherichia coli. The protein was purified in one chromatographic step after its in vitro activation. It was active on ABTS, 2,6-dimethoxyphenol (DMP), and anthraquinoid and azo dyes as reported for other fungal DyPs, but it was also able to oxidize Mn2+ (as manganese peroxidases and versatile peroxidases) and veratryl alcohol (VA) (as lignin peroxidases and versatile peroxidases). This corroborated that I. lacteus DyPs are the only enzymes able to oxidize high redox potential dyes, VA and Mn+2. Phylogenetic analysis grouped this enzyme with other type D-DyPs from basidiomycetes. In addition to its interest for dye decolorization, the results of the transformation of softwood and hardwood lignosulfonates suggest a putative biological role of this enzyme in the degradation of phenolic lignin.
Keywords: DyP; fungi; lignin; lignocellulosic biomass; oxidoreductases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Martínez Á.T., Speranza M., Ruiz-Dueñas F.J., Ferreira P., Camarero S., Guillén F., Martínez M.J., Gutiérrez A., Del Río J.C. Biodegradation of lignocellulosics: Microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 2005;8:195–204. - PubMed

