Multiregional Sequencing of IDH-WT Glioblastoma Reveals High Genetic Heterogeneity and a Dynamic Evolutionary History
- PMID: 33922652
- PMCID: PMC8122908
- DOI: 10.3390/cancers13092044
Multiregional Sequencing of IDH-WT Glioblastoma Reveals High Genetic Heterogeneity and a Dynamic Evolutionary History
Abstract
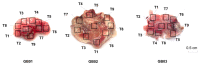
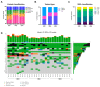
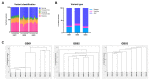
Glioblastoma is one of the most common and lethal primary neoplasms of the brain. Patient survival has not improved significantly over the past three decades and the patient median survival is just over one year. Tumor heterogeneity is thought to be a major determinant of therapeutic failure and a major reason for poor overall survival. This work aims to comprehensively define intra- and inter-tumor heterogeneity by mapping the genomic and mutational landscape of multiple areas of three primary IDH wild-type (IDH-WT) glioblastomas. Using whole exome sequencing, we explored how copy number variation, chromosomal and single loci amplifications/deletions, and mutational burden are spatially distributed across nine different tumor regions. The results show that all tumors exhibit a different signature despite the same diagnosis. Above all, a high inter-tumor heterogeneity emerges. The evolutionary dynamics of all identified mutations within each region underline the questionable value of a single biopsy and thus the therapeutic approach for the patient. Multiregional collection and subsequent sequencing are essential to try to address the clinical challenge of precision medicine. Especially in glioblastoma, this approach could provide powerful support to pathologists and oncologists in evaluating the diagnosis and defining the best treatment option.
Keywords: clonal evolution; glioblastoma; multiregional sequencing; spatial heterogeneity; temporal heterogeneity; tumor phylogeny; tumor progression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Gallaher J.A., Massey S.C., Hawkins-Daarud A., Noticewala S.S., Rockne R.C., Johnston S.K., Gonzalez-Cuyar L., Juliano J., Gil O., Swanson K.R., et al. From cells to tissue: How cell scale heterogeneity impacts glioblastoma growth and treatment response. PLoS Comput. Biol. 2020;16:e1007672. doi: 10.1371/journal.pcbi.1007672. - DOI - PMC - PubMed
-
- Mahlokozera T., Vellimana A.K., Li T., Mao D.D., Zohny Z.S., Kim D.H., Tran D.D., Marcus D.S., Fouke S.J., Campian J.L., et al. Biological and therapeutic implications of multisector sequencing in newly diagnosed glioblastoma. Neuro Oncol. 2018;20:472–483. doi: 10.1093/neuonc/nox232. - DOI - PMC - PubMed
-
- Sottoriva A., Spiteri I., Piccirillo S.G.M., Touloumis A., Collins V.P., Marioni J.C., Curtis C., Watts C., Tavaré S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. USA. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. - DOI - PMC - PubMed
-
- Weller M., van den Bent M., Tonn J.C., Stupp R., Preusser M., Cohen-Jonathan-Moyal E., Henriksson R., Le Rhun E., Balana C., Chinot O., et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18:e315–e329. doi: 10.1016/S1470-2045(17)30194-8. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

