Methoxy-Substituted Tyramine Derivatives Synthesis, Computational Studies and Tyrosinase Inhibitory Kinetics
- PMID: 33922836
- PMCID: PMC8122972
- DOI: 10.3390/molecules26092477
Methoxy-Substituted Tyramine Derivatives Synthesis, Computational Studies and Tyrosinase Inhibitory Kinetics
Abstract
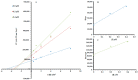
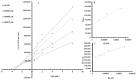
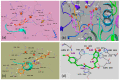
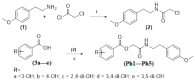
Targeting tyrosinase for melanogenesis disorders is an established strategy. Hydroxyl-substituted benzoic and cinnamic acid scaffolds were incorporated into new chemotypes that displayed in vitro inhibitory effects against mushroom and human tyrosinase for the purpose of identifying anti-melanogenic ingredients. The most active compound 2-((4-methoxyphenethyl)amino)-2-oxoethyl (E)-3-(2,4-dihydroxyphenyl) acrylate (Ph9), inhibited mushroom tyrosinase with an IC50 of 0.059 nM, while 2-((4-methoxyphenethyl)amino)-2-oxoethyl cinnamate (Ph6) had an IC50 of 2.1 nM compared to the positive control, kojic acid IC50 16700 nM. Results of human tyrosinase inhibitory activity in A375 human melanoma cells showed that compound (Ph9) and Ph6 exhibited 94.6% and 92.2% inhibitory activity respectively while the positive control kojic acid showed 72.9% inhibition. Enzyme kinetics reflected a mixed type of inhibition for inhibitor Ph9 (Ki 0.093 nM) and non-competitive inhibition for Ph6 (Ki 2.3 nM) revealed from Lineweaver-Burk plots. In silico docking studies with mushroom tyrosinase (PDB ID:2Y9X) predicted possible binding modes in the catalytic site for these active compounds. Ph9 displayed no PAINS (pan-assay interference compounds) alerts. Our results showed that compound Ph9 is a potential candidate for further development of tyrosinase inhibitors.
Keywords: computational studies; enzyme kinetics mechanism; inhibitory activity; tyramine derivatives; tyrosinase inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

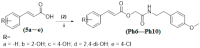
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

