Dieckol and Its Derivatives as Potential Inhibitors of SARS-CoV-2 Spike Protein (UK Strain: VUI 202012/01): A Computational Study
- PMID: 33922914
- PMCID: PMC8145291
- DOI: 10.3390/md19050242
Dieckol and Its Derivatives as Potential Inhibitors of SARS-CoV-2 Spike Protein (UK Strain: VUI 202012/01): A Computational Study
Abstract
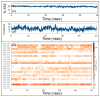
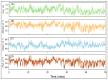
The high risk of morbidity and mortality associated with SARS-CoV-2 has accelerated the development of many potential vaccines. However, these vaccines are designed against SARS-CoV-2 isolated in Wuhan, China, and thereby may not be effective against other SARS-CoV-2 variants such as the United Kingdom variant (VUI-202012/01). The UK SARS-CoV-2 variant possesses D614G mutation in the Spike protein, which impart it a high rate of infection. Therefore, newer strategies are warranted to design novel vaccines and drug candidates specifically designed against the mutated forms of SARS-CoV-2. One such strategy is to target ACE2 (angiotensin-converting enzyme2)-Spike protein RBD (receptor binding domain) interaction. Here, we generated a homology model of Spike protein RBD of SARS-CoV-2 UK strain and screened a marine seaweed database employing different computational approaches. On the basis of high-throughput virtual screening, standard precision, and extra precision molecular docking, we identified BE011 (Dieckol) as the most potent compounds against RBD. However, Dieckol did not display drug-like properties, and thus different derivatives of it were generated in silico and evaluated for binding potential and drug-like properties. One Dieckol derivative (DK07) displayed good binding affinity for RBD along with acceptable physicochemical, pharmacokinetic, drug-likeness, and ADMET properties. Analysis of the RBD-DK07 interaction suggested the formation of hydrogen bonds, electrostatic interactions, and hydrophobic interactions with key residues mediating the ACE2-RBD interaction. Molecular dynamics simulation confirmed the stability of the RBD-DK07 complex. Free energy calculations suggested the primary role of electrostatic and Van der Waals' interaction in stabilizing the RBD-DK07 complex. Thus, DK07 may be developed as a potential inhibitor of the RBD-ACE2 interaction. However, these results warrant further validation by in vitro and in vivo studies.
Keywords: COVID-19; marine-derived compounds; molecular docking and simulation; natural compounds; seaweeds; spike protein.
Conflict of interest statement
All the authors declare that there are no competing interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

