S100P Interacts with p53 while Pentamidine Inhibits This Interaction
- PMID: 33923162
- PMCID: PMC8145327
- DOI: 10.3390/biom11050634
S100P Interacts with p53 while Pentamidine Inhibits This Interaction
Abstract
S100P, a small calcium-binding protein, associates with the p53 protein with micromolar affinity. It has been hypothesized that the oncogenic function of S100P may involve binding-induced inactivation of p53. We used 1H-15N HSQC experiments and molecular modeling to study the molecular interactions between S100P and p53 in the presence and absence of pentamidine. Our experimental analysis indicates that the S100P-53 complex formation is successfully disrupted by pentamidine, since S100P shares the same binding site for p53 and pentamidine. In addition, we showed that pentamidine treatment of ZR-75-1 breast cancer cells resulted in reduced proliferation and increased p53 and p21 protein levels, indicating that pentamidine is an effective antagonist that interferes with the S100P-p53 interaction, leading to re-activation of the p53-21 pathway and inhibition of cancer cell proliferation. Collectively, our findings suggest that blocking the association between S100P and p53 by pentamidine will prevent cancer progression and, therefore, provide a new avenue for cancer therapy by targeting the S100P-p53 interaction.
Keywords: 1H-5N HSQC spectrum; HADDOCK program; S100P; biomolecular docking; p53-TAD (73 amino acids); protein-protein interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

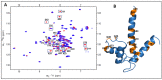


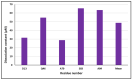


Similar articles
-
Pentamidine inhibit S100A4 - p53 interaction and decreases cell proliferation activity.Arch Biochem Biophys. 2020 Sep 30;691:108442. doi: 10.1016/j.abb.2020.108442. Epub 2020 Jul 7. Arch Biochem Biophys. 2020. PMID: 32649952
-
Structural insights into calcium-bound S100P and the V domain of the RAGE complex.PLoS One. 2014 Aug 1;9(8):e103947. doi: 10.1371/journal.pone.0103947. eCollection 2014. PLoS One. 2014. PMID: 25084534 Free PMC article.
-
Cancer-associated S100P protein binds and inactivates p53, permits therapy-induced senescence and supports chemoresistance.Oncotarget. 2016 Apr 19;7(16):22508-22. doi: 10.18632/oncotarget.7999. Oncotarget. 2016. PMID: 26967060 Free PMC article.
-
Transcriptional regulation and functional implication of S100P in cancer.Amino Acids. 2011 Oct;41(4):885-92. doi: 10.1007/s00726-010-0495-5. Epub 2010 Feb 14. Amino Acids. 2011. PMID: 20155429 Review.
-
S100P, a peculiar member of S100 family of calcium-binding proteins implicated in cancer.Acta Virol. 2013;57(2):238-46. doi: 10.4149/av_2013_02_238. Acta Virol. 2013. PMID: 23600880 Review.
Cited by
-
Repurposing the anti-parasitic agent pentamidine for cancer therapy; a novel approach with promising anti-tumor properties.J Transl Med. 2025 Mar 3;23(1):258. doi: 10.1186/s12967-025-06293-w. J Transl Med. 2025. PMID: 40033361 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

