Transcriptome Analysis of Seed Weight Plasticity in Brassica napus
- PMID: 33923211
- PMCID: PMC8123204
- DOI: 10.3390/ijms22094449
Transcriptome Analysis of Seed Weight Plasticity in Brassica napus
Abstract
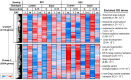
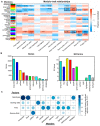
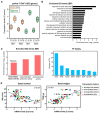
A critical barrier to improving crop yield is the trade-off between seed weight (SW) and seed number (SN), which has been commonly reported in several crops, including Brassica napus. Despite the agronomic relevance of this issue, the molecular factors involved in the interaction between SW and SN are largely unknown in crops. In this work, we performed a detailed transcriptomic analysis of 48 seed samples obtained from two rapeseed spring genotypes subjected to different source-sink (S-S) ratios in order to examine the relationship between SW and SN under different field conditions. A multifactorial analysis of the RNA-seq data was used to identify a group of 1014 genes exclusively regulated by the S-S ratio. We found that a reduction in the S-S ratio during seed filling induces the expression of genes involved in sucrose transport, seed weight, and stress responses. Moreover, we identified five co-expression modules that are positively correlated with SW and negatively correlated with SN. Interestingly, one of these modules was significantly enriched in transcription factors (TFs). Furthermore, our network analysis predicted several NAC TFs as major hubs underlying SW and SN compensation. Taken together, our study provides novel insights into the molecular factors associated with the SW-SN relationship in rapeseed and identifies TFs as potential targets when improving crop yield.
Keywords: Brassica napus; gene co-expression; network analysis; seed number; seed weight; source–sink; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- Savadi S. Molecular Regulation of Seed Development and Strategies for Engineering Seed Size in Crop Plants. Plant Growth Regul. 2018;84:401–422. doi: 10.1007/s10725-017-0355-3. - DOI
-
- Universidade Federal de Ouro Preto . UFOP Report on Global Market Supply 2017/2018. Universidade Federal de Ouro Preto; Ouro Preto, Brazil: 2019.
-
- FAO . Food Outlook—Biannual Report on Global Food Markets. FAO; Rome, Italy: 2020.
-
- FAOSTAT Crops Production 2021. [(accessed on 23 March 2021)];2021 Available online: http://www.fao.org/faostat/en/#data/QC.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

