Biochemical Characterization of the Amylase Activity from the New Haloarchaeal Strain Haloarcula sp. HS Isolated in the Odiel Marshlands
- PMID: 33923574
- PMCID: PMC8073556
- DOI: 10.3390/biology10040337
Biochemical Characterization of the Amylase Activity from the New Haloarchaeal Strain Haloarcula sp. HS Isolated in the Odiel Marshlands
Abstract
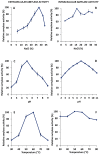
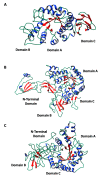
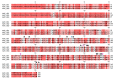
Alpha-amylases are a large family of α,1-4-endo-glycosyl hydrolases distributed in all kingdoms of life. The need for poly-extremotolerant amylases encouraged their search in extreme environments, where archaea become ideal candidates to provide new enzymes that are able to work in the harsh conditions demanded in many industrial applications. In this study, a collection of haloarchaea isolated from Odiel saltern ponds in the southwest of Spain was screened for their amylase activity. The strain that exhibited the highest activity was selected and identified as Haloarcula sp. HS. We demonstrated the existence in both, cellular and extracellular extracts of the new strain, of functional α-amylase activities, which showed to be moderately thermotolerant (optimum around 60 °C), extremely halotolerant (optimum over 25% NaCl), and calcium-dependent. The tryptic digestion followed by HPLC-MS/MS analysis of the partially purified cellular and extracellular extracts allowed to identify the sequence of three alpha-amylases, which despite sharing a low sequence identity, exhibited high three-dimensional structure homology, conserving the typical domains and most of the key consensus residues of α-amylases. Moreover, we proved the potential of the extracellular α-amylase from Haloarcula sp. HS to treat bakery wastes under high salinity conditions.
Keywords: amylase; enzymatic characterization; extremozymes; haloarchaea; proteomics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- Kanekar P.P., Kelkar A.S., Dhakephalkar P.K. Halophiles—Taxonomy, Diversity, Physiology and Applications. In: Metzler J.B., editor. Microorganisms in Environmental Management. Volume 9789400722. Springer; Dordrecht, The Netherlands: 2012. pp. 1–34.
-
- Ventosa A., Márquez M.C., Sánchez-Porro C., De La Haba R.R. Taxonomy of Halophilic Archaea and Bacteria. In: Vreeland R.H., editor. Advances in Understanding the Biology of Halophilic Microorganisms. Springer; Dordrecht, The Netherlands: 2012. pp. 59–80.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

