The Multifaceted Roles of Ku70/80
- PMID: 33923616
- PMCID: PMC8073936
- DOI: 10.3390/ijms22084134
The Multifaceted Roles of Ku70/80
Abstract
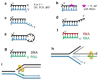
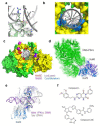
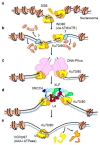
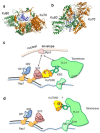
DNA double-strand breaks (DSBs) are accidental lesions generated by various endogenous or exogenous stresses. DSBs are also genetically programmed events during the V(D)J recombination process, meiosis, or other genome rearrangements, and they are intentionally generated to kill cancer during chemo- and radiotherapy. Most DSBs are processed in mammalian cells by the classical nonhomologous end-joining (c-NHEJ) pathway. Understanding the molecular basis of c-NHEJ has major outcomes in several fields, including radiobiology, cancer therapy, immune disease, and genome editing. The heterodimer Ku70/80 (Ku) is a central actor of the c-NHEJ as it rapidly recognizes broken DNA ends in the cell and protects them from nuclease activity. It subsequently recruits many c-NHEJ effectors, including nucleases, polymerases, and the DNA ligase 4 complex. Beyond its DNA repair function, Ku is also involved in several other DNA metabolism processes. Here, we review the structural and functional data on the DNA and RNA recognition properties of Ku implicated in DNA repair and in telomeres maintenance.
Keywords: DNA repair machinery; c-NHEJ; double-strand break; protein-DNA interactions; telomeres.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Mimori T., Akizuki M., Yamagata H., Inada S., Yoshida S., Homma M. Characterization of a high molecular weight acidic nuclear protein recognized by autoantibodies in sera from patients with polymyositis-scleroderma overlap. J. Clin. Investig. 1981;68:611–620. doi: 10.1172/JCI110295. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

