Current Methods of Post-Translational Modification Analysis and Their Applications in Blood Cancers
- PMID: 33923680
- PMCID: PMC8072572
- DOI: 10.3390/cancers13081930
Current Methods of Post-Translational Modification Analysis and Their Applications in Blood Cancers
Abstract
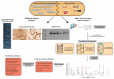
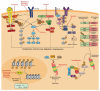
Post-translational modifications (PTMs) add a layer of complexity to the proteome through the addition of biochemical moieties to specific residues of proteins, altering their structure, function and/or localization. Mass spectrometry (MS)-based techniques are at the forefront of PTM analysis due to their ability to detect large numbers of modified proteins with a high level of sensitivity and specificity. The low stoichiometry of modified peptides means fractionation and enrichment techniques are often performed prior to MS to improve detection yields. Immuno-based techniques remain popular, with improvements in the quality of commercially available modification-specific antibodies facilitating the detection of modified proteins with high affinity. PTM-focused studies on blood cancers have provided information on altered cellular processes, including cell signaling, apoptosis and transcriptional regulation, that contribute to the malignant phenotype. Furthermore, the mechanism of action of many blood cancer therapies, such as kinase inhibitors, involves inhibiting or modulating protein modifications. Continued optimization of protocols and techniques for PTM analysis in blood cancer will undoubtedly lead to novel insights into mechanisms of malignant transformation, proliferation, and survival, in addition to the identification of novel biomarkers and therapeutic targets. This review discusses techniques used for PTM analysis and their applications in blood cancer research.
Keywords: acetylation; blood cancer; leukemia; lymphoma; multiple myeloma; myeloproliferative neoplasms; phosphorylation; post-translational modifications; sumoylation; ubiquitination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

