Antarctic Rahnella inusitata: A Producer of Cold-Stable β-Galactosidase Enzymes
- PMID: 33923711
- PMCID: PMC8074230
- DOI: 10.3390/ijms22084144
Antarctic Rahnella inusitata: A Producer of Cold-Stable β-Galactosidase Enzymes
Abstract
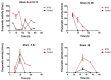
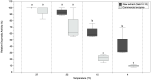
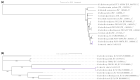
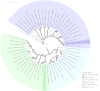
There has been a recent increase in the exploration of cold-active β-galactosidases, as it offers new alternatives for the dairy industry, mainly in response to the current needs of lactose-intolerant consumers. Since extremophilic microbial compounds might have unique physical and chemical properties, this research aimed to study the capacity of Antarctic bacterial strains to produce cold-active β-galactosidases. A screening revealed 81 out of 304 strains with β-galactosidase activity. The strain Se8.10.12 showed the highest enzymatic activity. Morphological, biochemical, and molecular characterization based on whole-genome sequencing confirmed it as the first Rahnella inusitata isolate from the Antarctic, which retained 41-62% of its β-galactosidase activity in the cold (4 °C-15 °C). Three β-galactosidases genes were found in the R. inusitata genome, which belong to the glycoside hydrolase families GH2 (LacZ and EbgA) and GH42 (BglY). Based on molecular docking, some of these enzymes exhibited higher lactose predicted affinity than the commercial control enzyme from Aspergillus oryzae. Hence, this work reports a new Rahnella inusitata strain from the Antarctic continent as a prominent cold-active β-galactosidase producer.
Keywords: Antarctica; cold-adapted bacteria; extremozymes; lactose; β-galactosidase.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Vásquez-Ponce F., Higuera-Llantén S., Pavlov M.S., Marshall S.H., Olivares-Pacheco J. Phylogenetic MLSA and phenotypic analysis identification of three probable novel Pseudomonas species isolated on King George Island, South Shetland, Antarctica. Braz. J. Microbiol. 2018;49:695–702. doi: 10.1016/j.bjm.2018.02.005. - DOI - PMC - PubMed
-
- Núñez-Montero K., Barrientos L. Advances in Antarctic Research for Antimicrobial Discovery: A Comprehensive Narrative Review of Bacteria from Antarctic Environments as Potential Sources of Novel Antibiotic Compounds Against Human Pathogens and Microorganisms of Industrial Importance. Antibiotics. 2018;7:90. doi: 10.3390/antibiotics7040090. - DOI - PMC - PubMed
-
- Nichols D., Bowman J., Sanderson K., Nichols C.M., Lewis T., McMeekin T., Nichols P.D. Developments with Antarctic microorganisms: Culture collections, bioactivity screening, taxonomy, PUFA production and cold-adapted enzymes. Curr. Opin. Biotechnol. 1999;10:240–246. doi: 10.1016/S0958-1669(99)80042-1. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- INACH RT_14‒12 and INACH DG_01-19/INSTITUTO ANTÁRTICO CHILENO (INACH)
- INI4/UFRO/UANDES
- DI19-0079/UNIVERSIDAD DE LA FRONTERA
- NXR17-0003/NETWORK FOR EXTREME ENVIRONMENTS RESEARCH (NEXER)
- CONICYT‒PFCHA/Doctorado Nacional/2017‒21170263 for K.N-M and CONICYT‒PFCHA/Doctorado Nacional/ 2017‒21171392/ANID (NATIONAL INVESTIGATION AND DEVELOPMENT AGENCY, CHILE)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

