Computational and Transcriptomic Analysis Unraveled OsMATE34 as a Putative Anthocyanin Transporter in Black Rice (Oryza sativa L.) Caryopsis
- PMID: 33923742
- PMCID: PMC8073145
- DOI: 10.3390/genes12040583
Computational and Transcriptomic Analysis Unraveled OsMATE34 as a Putative Anthocyanin Transporter in Black Rice (Oryza sativa L.) Caryopsis
Abstract
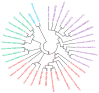
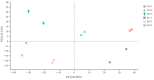
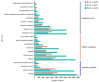
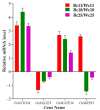
Anthocyanin is a flavonoid compound with potential antioxidant properties beneficial to human health and sustains plant growth and development under different environmental stresses. In black rice, anthocyanin can be found in the stems, leaves, stigmas, and caryopsis. Although the anthocyanin biosynthesis in rice has been extensively studied, limited knowledge underlying the storage mechanism and transporters is available. This study undertook the complementation of computational and transcriptome analysis to decipher a potential multidrug and toxic compound extrusion (MATE) gene candidate for anthocyanin transportation in black rice caryopsis. The phylogenetic analysis showed that OsMATE34 has the same evolutionary history and high similarities with VvAM1, VvAM3, MtMATE2, SlMATE/MTP77, RsMATE8, AtFFT, and AtTT12 involved in anthocyanin transportation. RNA sequencing analysis in black caryopsis (Bc; Bc11, Bc18, Bc25) and white caryopsis (Wc; Wc11, Wc18, Wc25), respectively, at 11 days after flowering (DAF), 18 DAF, and 25 DAF revealed a total of 36,079 expressed genes, including 33,157 known genes and 2922 new genes. The differentially expressed genes (DEGs) showed 15,573 genes commonly expressed, with 1804 and 1412 genes uniquely expressed in Bc and Wc, respectively. Pairwise comparisons showed 821 uniquely expressed genes out of 15,272 DEGs for Wc11 vs. Bc11, 201 uniquely expressed genes out of 16,240 DEGs for Wc18 vs. Bc18, and 2263 uniquely expressed genes out of 16,240 DEGs for Wc25 vs. Bc25. Along with anthocyanin biosynthesis genes (OsPAL, OsCHS, OsCHI, OsF3H, OsDFR, OsANS, and OsUFGT/Os3GT), OsMATE34 expression was significantly upregulated in all Bc but not in Wc. OsMATE34 expression was similar to OsGSTU34, a transporter of anthocyanin in rice leaves. Taken together, our results highlighted OsMATE34 (Os08g0562800) as a candidate anthocyanin transporter in rice caryopsis. This study provides a new finding and a clue to enhance the accumulation of anthocyanin in rice caryopsis.
Keywords: MATE transporters; anthoMATE; anthocyanin; anthocyanin’s transport mechanism; antioxidant; black rice caryopsis; cyanidin-3-glucoside; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Integrative HPLC profiling and transcriptome analysis revealed insights into anthocyanin accumulation and key genes at three developmental stages of black rice (Oryza sativa. L) caryopsis.Front Plant Sci. 2023 Aug 30;14:1211326. doi: 10.3389/fpls.2023.1211326. eCollection 2023. Front Plant Sci. 2023. PMID: 37727854 Free PMC article.
-
OsGSTU34, a Bz2-like anthocyanin-related glutathione transferase transporter, is essential for rice (Oryza sativa L.) organs coloration.Phytochemistry. 2024 Jan;217:113896. doi: 10.1016/j.phytochem.2023.113896. Epub 2023 Oct 20. Phytochemistry. 2024. PMID: 37866445
-
Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves.New Phytol. 2019 Jul;223(2):705-721. doi: 10.1111/nph.15807. Epub 2019 Apr 23. New Phytol. 2019. PMID: 30891753
-
Recent Insights into Anthocyanin Pigmentation, Synthesis, Trafficking, and Regulatory Mechanisms in Rice (Oryza sativa L.) Caryopsis.Biomolecules. 2021 Mar 7;11(3):394. doi: 10.3390/biom11030394. Biomolecules. 2021. PMID: 33800105 Free PMC article. Review.
-
Recent insights into anthocyanin biosynthesis, gene involvement, distribution regulation, and domestication process in rice (Oryza sativa L.).Plant Sci. 2024 Dec;349:112282. doi: 10.1016/j.plantsci.2024.112282. Epub 2024 Oct 9. Plant Sci. 2024. PMID: 39389316 Review.
Cited by
-
Integrative HPLC profiling and transcriptome analysis revealed insights into anthocyanin accumulation and key genes at three developmental stages of black rice (Oryza sativa. L) caryopsis.Front Plant Sci. 2023 Aug 30;14:1211326. doi: 10.3389/fpls.2023.1211326. eCollection 2023. Front Plant Sci. 2023. PMID: 37727854 Free PMC article.
-
Advances and Future Prospects of Pigment Deposition in Pigmented Rice.Plants (Basel). 2025 Mar 19;14(6):963. doi: 10.3390/plants14060963. Plants (Basel). 2025. PMID: 40265906 Free PMC article. Review.
-
Rice co-expression network analysis identifies gene modules associated with agronomic traits.Plant Physiol. 2022 Sep 28;190(2):1526-1542. doi: 10.1093/plphys/kiac339. Plant Physiol. 2022. PMID: 35866684 Free PMC article.
-
Genome-Wide Association Study Reveals the Genetic Basis of Total Flavonoid Content in Brown Rice.Genes (Basel). 2023 Aug 25;14(9):1684. doi: 10.3390/genes14091684. Genes (Basel). 2023. PMID: 37761824 Free PMC article.
-
Plant Secondary Metabolite Transporters: Diversity, Functionality, and Their Modulation.Front Plant Sci. 2021 Oct 27;12:758202. doi: 10.3389/fpls.2021.758202. eCollection 2021. Front Plant Sci. 2021. PMID: 34777438 Free PMC article. Review.
References
-
- Ali I., He L., Ullah S., Quan Z., Wei S., Iqbal A., Munsif F., Shah T., Xuan Y., Luo Y., et al. Biochar addition coupled with nitrogen fertilization impacts on soil quality, crop productivity, and nitrogen uptake under double-cropping system. Food Energy Secur. 2020;9:e208. doi: 10.1002/fes3.208. - DOI
-
- Ranganathan J., Waite R., Searchinger T., Hanson C. How to Sustainably Feed 10 Billion People by 2050, in 21 Charts. [(accessed on 10 April 2020)];World Res. Ins. 2018 Available online: https://www.wri.org/blog/2018/12/how-sustainably-feed-10-billion-people-....
-
- Terungwa T.I., Yuguda M. The impact of rice production, consumption and importation in Nigeria: The political economy perspectives. Int. J. Sustain. Dev. World Policy. 2014;3:90–99.
-
- Hao J., Zhu H., Zhang Z., Yang S., Li H. Identification of anthocyanins in black rice (Oryza sativa L.) by UPLC/Q-TOF-MS and their in-vitro and in-vivo antioxidant activities. J. Cereal Sci. 2015;64:92–99. doi: 10.1016/j.jcs.2015.05.003. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

