A Remarkable Genetic Diversity of Rotavirus A Circulating in Red Fox Population in Croatia
- PMID: 33923799
- PMCID: PMC8072941
- DOI: 10.3390/pathogens10040485
A Remarkable Genetic Diversity of Rotavirus A Circulating in Red Fox Population in Croatia
Abstract
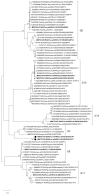
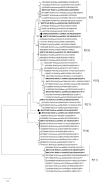
Rotaviruses (RV), especially Rotavirus A (RVA), are globally recognized as pathogens causing neonatal diarrhea, but they also affect intensive animal farming. However, the knowledge on their significance in wildlife is rather limited. The aim of the study was to unveil the prevalence, molecular epidemiology, and genetic diversity of RVA strains circulating in the red fox (Vulpes vulpes) population in Croatia. From 2018 to 2019, 370 fecal samples from fox carcasses hunted for rabies monitoring were collected. All samples were first tested using a VP2 real-time RT-PCR; in the subsequent course, positives were subjected to VP7 and VP4 genotyping. The results revealed an RVA prevalence of 14.9%, while the circulating RVA strains showed a remarkable genetic diversity in terms of 11 G and nine P genotypes, among which one G and three P were tentatively identified as novel. In total, eight genotype combinations were detected: G8P[14], G9P[3], G9P[23], G10P[11], G10P[3], G11P[13], G15P[21], and G?P[?]. The results suggest a complex background of previous interspecies transmission events, shedding new light on the potential influence of foxes in RVA epidemiology. Their role as potential reservoirs of broad range of RVA genotypes, usually considered typical solely of domestic animals and humans, cannot be dismissed.
Keywords: Croatia; Rotavirus A; genetic diversity; genotype; interspecies transmission; molecular epidemiology; phylogenetic analysis; red fox.
Conflict of interest statement
The authors declare no conflict of interest. The funding body had no role in the study design; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the study.
Figures

References
-
- Troeger C., Khalil I.A., Rao P.C., Cao S., Blacker B.F., Ahmed T., Armah G., Bines J.E., Brewer T.G., Colombara D.V., et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018;172:958–965. doi: 10.1001/jamapediatrics.2018.1960. - DOI - PMC - PubMed
-
- Hungerford D., Vivancos R., Read J.M., Pitzer V.E., Cunliffe N., French N., Iturriza-Gomara M. In-season and out-of-season variation of rotavirus genotype distribution and age of infection across 12 European countries before the introduction of routine vaccination, 2007/08 to 2012/13. Eurosurveillance. 2016;21:21. doi: 10.2807/1560-7917.ES.2016.21.2.30106. - DOI - PubMed
-
- Palmarini M. Reoviridae. In: MacLachlan N.J., Dubovi E.J., editors. Fenner’s Veterinary Virology. 5th ed. Academic Press; Boston, MA, USA: 2017. pp. 299–317.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

