Comparing Sediment Microbiomes in Contaminated and Pristine Wetlands along the Coast of Yucatan
- PMID: 33923859
- PMCID: PMC8073884
- DOI: 10.3390/microorganisms9040877
Comparing Sediment Microbiomes in Contaminated and Pristine Wetlands along the Coast of Yucatan
Abstract
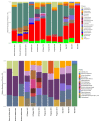
Microbial communities are important players in coastal sediments for the functioning of the ecosystem and the regulation of biogeochemical cycles. They also have great potential as indicators of environmental perturbations. To assess how microbial communities can change their composition and abundance along coastal areas, we analyzed the composition of the microbiome of four locations of the Yucatan Peninsula using 16S rRNA gene amplicon sequencing. To this end, sediment from two conserved (El Palmar and Bocas de Dzilam) and two contaminated locations (Sisal and Progreso) from the coast northwest of the Yucatan Peninsula in three different years, 2017, 2018 and 2019, were sampled and sequenced. Microbial communities were found to be significantly different between the locations. The most noticeable difference was the greater relative abundance of Planctomycetes present at the conserved locations, versus FBP group found with greater abundance in contaminated locations. In addition to the difference in taxonomic groups composition, there is a variation in evenness, which results in the samples of Bocas de Dzilam and Progreso being grouped separately from those obtained in El Palmar and Sisal. We also carry out the functional prediction of the metabolic capacities of the microbial communities analyzed, identifying differences in their functional profiles. Our results indicate that landscape of the coastal microbiome of Yucatan sediment shows changes along the coastline, reflecting the constant dynamics of coastal environments and their impact on microbial diversity.
Keywords: Yucatan Peninsula; coastal prokaryotic communities; metagenomic.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






References
-
- Quero G.M., Cassin D., Botter M., Perini L., Luna G.M. Patterns of benthic bacterial diversity in coastal areas contaminated by heavy metals, polycyclic aromatic hydrocarbons (PAHs) and polychlorinated biphenyls (PCBs) Front. Microbiol. 2015;6:1053. doi: 10.3389/fmicb.2015.01053. - DOI - PMC - PubMed
-
- Wang Y., Sheng H.-F., He Y., Wu J.-Y., Jiang Y.-X., Tam N.F.-Y., Zhou H.-W. Comparison of the Levels of Bacterial Diversity in Freshwater, Intertidal Wetland, and Marine Sediments by Using Millions of Illumina Tags. Appl. Environ. Microbiol. 2012;78:8264–8271. doi: 10.1128/AEM.01821-12. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

