Streamlined Multimodal DESI and MALDI Mass Spectrometry Imaging on a Singular Dual-Source FT-ICR Mass Spectrometer
- PMID: 33923908
- PMCID: PMC8073082
- DOI: 10.3390/metabo11040253
Streamlined Multimodal DESI and MALDI Mass Spectrometry Imaging on a Singular Dual-Source FT-ICR Mass Spectrometer
Abstract
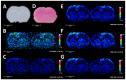
The study of biological specimens by mass spectrometry imaging (MSI) has had a profound influence in the various forms of spatial-omics over the past two decades including applications for the identification of clinical biomarker analysis; the metabolic fingerprinting of disease states; treatment with therapeutics; and the profiling of lipids, peptides and proteins. No singular approach is able to globally map all biomolecular classes simultaneously. This led to the development of many complementary multimodal imaging approaches to solve analytical problems: fusing multiple ionization techniques, imaging microscopy or spectroscopy, or local extractions into robust multimodal imaging methods. However, each fusion typically requires the melding of analytical information from multiple commercial platforms, and the tandem utilization of multiple commercial or third-party software platforms-even in some cases requiring computer coding. Herein, we report the use of matrix-assisted laser desorption/ionization (MALDI) in tandem with desorption electrospray ionization (DESI) imaging in the positive ion mode on a singular commercial orthogonal dual-source Fourier transform ion cyclotron resonance (FT-ICR) instrument for the complementary detection of multiple analyte classes by MSI from tissue. The DESI source was 3D printed and the commercial Bruker Daltonics software suite was used to generate mass spectrometry images in tandem with the commercial MALDI source. This approach allows for the generation of multiple modes of mass spectrometry images without the need for third-party software and a customizable platform for ambient ionization imaging. Highlighted is the streamlined workflow needed to obtain phospholipid profiles, as well as increased depth of coverage of both annotated phospholipid, cardiolipin, and ganglioside species from rat brain with both high spatial and mass resolution.
Keywords: 3D printed/printing; Fourier transform ion cyclotron resonance (FT-ICR); desorption electrospray ionization (DESI); lipidomics; mass spectrometry imaging (MSI); matrix-assisted laser desorption/ionization (MALDI); multimodal imaging.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- Wiseman J.M., Ifa D.R., Zhu Y., Kissinger C.B., Nicholas E., Manicke N.E., Peter T., Kissinger P.T., Cooks R.G. Desorption Electrospray Ionization Mass Spectrometry: Imaging Drugs and Metabolites in Tissues. Proc. Natl. Acad. Sci. USA. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

