Characterization of Recombinant Adeno-Associated Viruses (rAAVs) for Gene Therapy Using Orthogonal Techniques
- PMID: 33923984
- PMCID: PMC8074050
- DOI: 10.3390/pharmaceutics13040586
Characterization of Recombinant Adeno-Associated Viruses (rAAVs) for Gene Therapy Using Orthogonal Techniques
Abstract
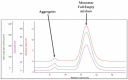
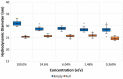
Viruses are increasingly used as vectors for delivery of genetic material for gene therapy and vaccine applications. Recombinant adeno-associated viruses (rAAVs) are a class of viral vector that is being investigated intensively in the development of gene therapies. To develop efficient rAAV therapies produced through controlled and economical manufacturing processes, multiple challenges need to be addressed starting from viral capsid design through identification of optimal process and formulation conditions to comprehensive quality control. Addressing these challenges requires fit-for-purpose analytics for extensive characterization of rAAV samples including measurements of capsid or particle titer, percentage of full rAAV particles, particle size, aggregate formation, thermal stability, genome release, and capsid charge, all of which may impact critical quality attributes of the final product. Importantly, there is a need for rapid analytical solutions not relying on the use of dedicated reagents and costly reference standards. In this study, we evaluate the capabilities of dynamic light scattering, multiangle dynamic light scattering, and SEC-MALS for analyses of rAAV5 samples in a broad range of viral concentrations (titers) at different levels of genome loading, sample heterogeneity, and sample conditions. The study shows that DLS and MADLS® can be used to determine the size of full and empty rAAV5 (27 ± 0.3 and 33 ± 0.4 nm, respectively). A linear range for rAAV5 size and titer determination with MADLS was established to be 4.4 × 1011-8.7 × 1013 cp/mL for the nominally full rAAV5 samples and 3.4 × 1011-7 × 1013 cp/mL for the nominally empty rAAV5 samples with 3-8% and 10-37% CV for the full and empty rAAV5 samples, respectively. The structural stability and viral load release were also inferred from a combination of DLS, SEC-MALS, and DSC. The structural characteristics of the rAAV5 start to change from 40 °C onward, with increasing aggregation observed. With this study, we explored and demonstrated the applicability and value of orthogonal and complementary label-free technologies for enhanced serotype-independent characterization of key properties and stability profiles of rAAV5 samples.
Keywords: SEC–MALS; differential scanning calorimetry (DSC); dynamic light scattering (DLS); high-throughput; multiangle dynamic light scattering (MADLS®); orthogonal techniques; recombinant adeno-associated viruses (rAAV).
Conflict of interest statement
The company had a role in manufacturing technologies used in the paper. The authors declare no conflict of interest.
Figures










References
-
- Singh P., Gonzalez M.J., Manchester M. Viruses and their uses in nanotechnology. Drug Dev. Res. 2006;67:23–41. doi: 10.1002/ddr.20064. - DOI
-
- Manchester M., Steinmetz N.F. Viruses and Nanotechnology. Springer; Berlin/Heidelberg, Germany: 2008.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

