Global Discrepancies between Numbers of Available SARS-CoV-2 Genomes and Human Development Indexes at Country Scales
- PMID: 33924778
- PMCID: PMC8145975
- DOI: 10.3390/v13050775
Global Discrepancies between Numbers of Available SARS-CoV-2 Genomes and Human Development Indexes at Country Scales
Abstract
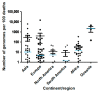
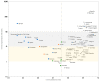
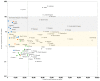
It has now been over a year since SARS-CoV-2 first emerged in China, in December 2019, and it has spread rapidly around the world. Some variants are currently considered of great concern. We aimed to analyze the numbers of SARS-CoV-2 genome sequences obtained in different countries worldwide until January 2021. On 28 January 2021, we downloaded the deposited genome sequence origin from the GISAID database, and from the "Our world in data" website we downloaded numbers of SARS-CoV-2-diagnosed cases, numbers of SARS-CoV-2-associated deaths, population size, life expectancy, gross domestic product (GDP) per capita, and human development index per country. Files were merged and data were analyzed using Microsoft Excel software. A total of 450,968 SARS-CoV-2 genomes originating from 135 countries on the 5 continents were available. When considering the 19 countries for which the number of genomes per 100 deaths was >100, six were in Europe, while eight were in Asia, three were in Oceania and two were in Africa. Six (30%) of these countries are beyond rank 75, regarding the human development index and four (20%) are beyond rank 80 regarding GDP per capita. Moreover, the comparisons of the number of genomes sequenced per 100 deaths to the human development index by country show that some Western European countries have released similar or lower numbers of genomes than many African or Asian countries with a lower human development index. Previous data highlight great discrepancies between the numbers of available SARS-CoV-2 genomes per 100 cases and deaths and the ranking of countries regarding wealth and development.
Keywords: SARS-CoV-2; country-scale development; genome; next-generation sequencing; variant; world.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Levasseur A., Delerce J., Caputo A., Brechard L., Colson P., Lagier J.C., Fournier P.E., Raoult D. Genomic diversity and evolution of coronavirus (SARS-CoV-2) in France from 309 COVID-19-infected patients. bioRxiv. 2020 doi: 10.1101/2020.09.04.282616. - DOI
-
- Colson P., Levasseur A., Delerce J., Chaudet H., Bossi V., Ben Khedher M., Fournier P.E., Lagier J.C., Raoult D. Dramatic increase in the SARS-CoV-2 mutation rate and low mortality rate during the second epidemic in summer in Marseille. IHU Pre-Prints. 2020 doi: 10.35088/68c3-ew82. - DOI
-
- Colson P., Levasseur A., Gautret P., Fenollar F., Thuan H.V., Delerce J., Bitam I., Saile R., Maaloum M., Padane A., et al. Introduction into the Marseille geographical area of a mild SARS-CoV-2 variant originating from sub-Saharan Africa: An investigational study. Travel Med. Infect. Dis. 2021;40:101980. doi: 10.1016/j.tmaid.2021.101980. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

