CRISPR Interference Efficiently Silences Latent and Lytic Viral Genes in Kaposi's Sarcoma-Associated Herpesvirus-Infected Cells
- PMID: 33924938
- PMCID: PMC8146339
- DOI: 10.3390/v13050783
CRISPR Interference Efficiently Silences Latent and Lytic Viral Genes in Kaposi's Sarcoma-Associated Herpesvirus-Infected Cells
Abstract
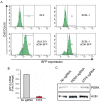
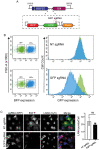
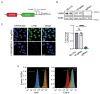
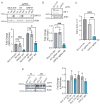
Uncovering viral gene functions requires the modulation of gene expression through overexpression or loss-of-function. CRISPR interference (CRISPRi), a modification of the CRISPR-Cas9 gene editing technology, allows specific and efficient transcriptional silencing without genetic ablation. CRISPRi has been used to silence eukaryotic and prokaryotic genes at the single-gene and genome-wide levels. Here, we report the use of CRISPRi to silence latent and lytic viral genes, with an efficiency of ~80-90%, in epithelial and B-cells carrying multiple copies of the Kaposi's sarcoma-associated herpesvirus (KSHV) genome. Our results validate CRISPRi for the analysis of KSHV viral elements, providing a functional genomics tool for studying virus-host interactions.
Keywords: CRISPR-interference; KSHV; Kaposi’s sarcoma-associated herpesvirus; dCas9-KRAB; gene expression; gene silencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- International Agency for Research on Cancer . Globocan 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. World Health Organisation (WHO); Geneva, Switzerland: 2015.
-
- What Are the Key Statistics about Kaposi Sarcoma? American Cancer Society; Burlington, VT, USA: 2016.
-
- Arias C., Weisburd B., Stern-Ginossar N., Mercier A., Madrid A.S., Bellare P., Holdorf M., Weissman J.S., Ganem D. KSHV 2.0: A Comprehensive Annotation of the Kaposi’s Sarcoma-Associated Herpesvirus Genome Using Next-Generation Sequencing Reveals Novel Genomic and Functional Features. PLoS Pathog. 2014;10:e1003847. doi: 10.1371/journal.ppat.1003847. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

