Identification of CRYAB+ KCNN3+ SOX9+ Astrocyte-Like and EGFR+ PDGFRA+ OLIG1+ Oligodendrocyte-Like Tumoral Cells in Diffuse IDH1-Mutant Gliomas and Implication of NOTCH1 Signalling in Their Genesis
- PMID: 33925547
- PMCID: PMC8123787
- DOI: 10.3390/cancers13092107
Identification of CRYAB+ KCNN3+ SOX9+ Astrocyte-Like and EGFR+ PDGFRA+ OLIG1+ Oligodendrocyte-Like Tumoral Cells in Diffuse IDH1-Mutant Gliomas and Implication of NOTCH1 Signalling in Their Genesis
Abstract
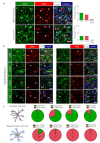
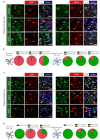
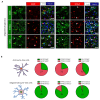
Diffuse grade II IDH-mutant gliomas are slow-growing brain tumors that progress into high-grade gliomas. They present intratumoral cell heterogeneity, and no reliable markers are available to distinguish the different cell subtypes. The molecular mechanisms underlying the formation of this cell diversity is also ill-defined. Here, we report that SOX9 and OLIG1 transcription factors, which specifically label astrocytes and oligodendrocytes in the normal brain, revealed the presence of two largely nonoverlapping tumoral populations in IDH1-mutant oligodendrogliomas and astrocytomas. Astrocyte-like SOX9+ cells additionally stained for APOE, CRYAB, ID4, KCNN3, while oligodendrocyte-like OLIG1+ cells stained for ASCL1, EGFR, IDH1, PDGFRA, PTPRZ1, SOX4, and SOX8. GPR17, an oligodendrocytic marker, was expressed by both cells. These two subpopulations appear to have distinct BMP, NOTCH1, and MAPK active pathways as stainings for BMP4, HEY1, HEY2, p-SMAD1/5 and p-ERK were higher in SOX9+ cells. We used primary cultures and a new cell line to explore the influence of NOTCH1 activation and BMP treatment on the IDH1-mutant glioma cell phenotype. This revealed that NOTCH1 globally reduced oligodendrocytic markers and IDH1 expression while upregulating APOE, CRYAB, HEY1/2, and an electrophysiologically-active Ca2+-activated apamin-sensitive K+ channel (KCNN3/SK3). This was accompanied by a reduction in proliferation. Similar effects of NOTCH1 activation were observed in nontumoral human oligodendrocytic cells, which additionally induced strong SOX9 expression. BMP treatment reduced OLIG1/2 expression and strongly upregulated CRYAB and NOGGIN, a negative regulator of BMP. The presence of astrocyte-like SOX9+ and oligodendrocyte-like OLIG1+ cells in grade II IDH1-mutant gliomas raises new questions about their role in the pathology.
Keywords: BMP; NOTCH1 pathway; brain tumors; cellular heterogeneity; diffuse IDH1-mutant gliomas; diffuse grade II IDH-mutant glioma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Louis D.N., Ohgaki H., Wiestler O.D., Cavenee W.K. World Health Organization Histological Classification of Tumours of the Central Nervous System. International Agency for Research on Cancer; Lyon, France: 2016.
-
- Pekmezci M., Rice T., Molinaro A.M., Walsh K.M., Decker P.A., Hansen H., Sicotte H., Kollmeyer T.M., McCoy L.S., Sarkar G., et al. Adult infiltrating gliomas with WHO 2016 integrated diagnosis: Additional prognostic roles of ATRX and TERT. Acta Neuropathol. 2017;133:1001–1016. doi: 10.1007/s00401-017-1690-1. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

