Estimating Genetic Similarity Matrices Using Phylogenies
- PMID: 33926225
- PMCID: PMC8219189
- DOI: 10.1089/cmb.2020.0375
Estimating Genetic Similarity Matrices Using Phylogenies
Abstract
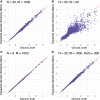
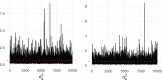
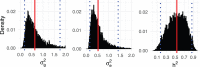
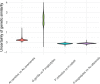
Genetic similarity is a measure of the genetic relatedness among individuals. The standard method for computing these matrices involves the inner product of observed genetic variants. Such an approach is inaccurate or impossible if genotypes are not available, or not densely sampled, or of poor quality (e.g., genetic analysis of extinct species). We provide a new method for computing genetic similarities among individuals using phylogenetic trees. Our method can supplement (or stand in for) computations based on genotypes. We provide simulations suggesting that the genetic similarity matrices computed from trees are consistent with those computed from genotypes. With our methods, quantitative analysis on genetic traits and analysis of heritability and coheritability can be conducted directly using genetic similarity matrices and so in the absence of genotype data, or under uncertainty in the phylogenetic tree. We use simulation studies to demonstrate the advantages of our method, and we provide applications to data.
Keywords: genetic similarity; infinite sites model; phylogenetic tree.
Conflict of interest statement
No competing financial interests exist.
Figures




References
-
- Atkinson, Q.D., and Gray, R.D.. 2005. Curious parallels and curious connections—phylogenetic thinking in biology and historical linguistics. Syst. Biol. 54, 513–526 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
