Probabilistic Atlas of the Mesencephalic Reticular Formation, Isthmic Reticular Formation, Microcellular Tegmental Nucleus, Ventral Tegmental Area Nucleus Complex, and Caudal-Rostral Linear Raphe Nucleus Complex in Living Humans from 7 Tesla Magnetic Resonance Imaging
- PMID: 33926237
- PMCID: PMC8817713
- DOI: 10.1089/brain.2020.0975
Probabilistic Atlas of the Mesencephalic Reticular Formation, Isthmic Reticular Formation, Microcellular Tegmental Nucleus, Ventral Tegmental Area Nucleus Complex, and Caudal-Rostral Linear Raphe Nucleus Complex in Living Humans from 7 Tesla Magnetic Resonance Imaging
Abstract
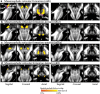
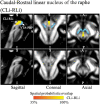
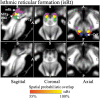
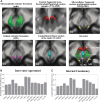
Introduction: The mesencephalic reticular formation, isthmic reticular formation, microcellular tegmental nucleus, ventral tegmental area-parabrachial pigmented nucleus complex, and caudal-rostral linear nucleus of the raphe are small brainstem regions crucially involved in arousal, sleep, and reward. Yet, these nuclei are difficult to identify with magnetic resonance imaging (MRI) of living humans. In the current work, we developed a probabilistic atlas of these brainstem nuclei in living humans, using noninvasive ultra-high-field MRI. Methods: We acquired single-subject, multicontrast (diffusion and T2-weighted), 1.1-mm isotropic resolution, 7 Tesla MRI images of 12 healthy subjects. After preprocessing and alignment to the stereotactic space, these images were used to delineate (in each subject) the nuclei of interest based on the image contrast as well as on neighboring nuclei and landmarks. Nucleus labels were averaged across subjects to yield probabilistic labels. The latter were further validated by assessment of the label inter-rater agreement, internal consistency, and volume. Results: Labels were delineated for each nucleus with good overlap across subjects. The inter-rater agreement and internal consistency were below (p < 10-8) the linear spatial imaging resolution (1.1 mm), thus validating the generated probabilistic atlas labels. The volumes of our labels did not differ from literature volumes (p < 0.05), further validating our atlas. Discussion and Conclusion: The probabilistic atlas of these five mesopontine nuclei expands current in vivo brainstem nuclei atlases and can be used as a tool to identify the location of these areas in conventional (e.g., 3 Tesla) images. This might serve to unravel the brainstem structure-to-function link and thus improve clinical outcomes. Impact statement The mesencephalic reticular formation, isthmic reticular formation, microcellular tegmental nucleus, ventral tegmental area-parabrachial pigmented nucleus complex, and caudal-rostral linear nucleus of the raphe are small brainstem regions crucially involved in arousal, sleep, and reward. In the current work, we developed a probabilistic atlas of these brainstem nuclei in living humans, using noninvasive, ultra-high-field magnetic resonance imaging. The probabilistic atlas of these five mesopontine nuclei expands current in vivo brainstem nuclei atlases and can be used as a tool to identify the location of these areas in conventional (e.g., 3 Tesla) images. This might serve to unravel the brainstem structure-to-function link and thus improve clinical outcomes.
Keywords: brainstem nuclei atlas; caudal-rostral linear nucleus of the raphe; isthmic reticular formation; mesencephalic reticular formation; microcellular tegmental nucleus; ventral tegmental area-parabrachial pigmented nucleus complex.
Conflict of interest statement
Authors have no conflicts of interest to disclose.
Figures


References
-
- Ayachit U. 2015. The Paraview Guide: A Parallel Visualization Application. NY, USA: Kitware, Inc.
-
- Barreiro-Iglesias A, Villar-Cerviño V, Anadón R, et al. 2008. Development and organization of the descending serotonergic brainstem-spinal projections in the sea lamprey. J Chem Neuroanat 36:77–84. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

