Genome-wide association study identifies RNF123 locus as associated with chronic widespread musculoskeletal pain
- PMID: 33926923
- PMCID: PMC8372387
- DOI: 10.1136/annrheumdis-2020-219624
Genome-wide association study identifies RNF123 locus as associated with chronic widespread musculoskeletal pain
Abstract
Background and objectives: Chronic widespread musculoskeletal pain (CWP) is a symptom of fibromyalgia and a complex trait with poorly understood pathogenesis. CWP is heritable (48%-54%), but its genetic architecture is unknown and candidate gene studies have produced inconsistent results. We conducted a genome-wide association study to get insight into the genetic background of CWP.
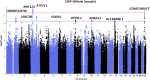
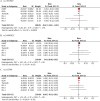
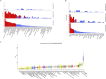
Methods: Northern Europeans from UK Biobank comprising 6914 cases reporting pain all over the body lasting >3 months and 242 929 controls were studied. Replication of three independent genome-wide significant single nucleotide polymorphisms was attempted in six independent European cohorts (n=43 080; cases=14 177). Genetic correlations with risk factors, tissue specificity and colocalisation were examined.
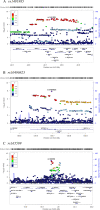
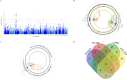
Results: Three genome-wide significant loci were identified (rs1491985, rs10490825, rs165599) residing within the genes Ring Finger Protein 123 (RNF123), ATPase secretory pathway Ca2+transporting 1 (ATP2C1) and catechol-O-methyltransferase (COMT). The RNF123 locus was replicated (meta-analysis p=0.0002), the ATP2C1 locus showed suggestive association (p=0.0227) and the COMT locus was not replicated. Partial genetic correlation between CWP and depressive symptoms, body mass index, age of first birth and years of schooling were identified. Tissue specificity and colocalisation analysis highlight the relevance of skeletal muscle in CWP.
Conclusions: We report a novel association of RNF123 locus and a suggestive association of ATP2C1 locus with CWP. Both loci are consistent with a role of calcium regulation in CWP. The association with COMT, one of the most studied genes in chronic pain field, was not confirmed in the replication analysis.
Keywords: epidemiology; fibromyalgia; genetic; polymorphism.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: YSA is co-owner of Maatschap PolyOmica and PolyKnomics BV, private organisations, providing services, research and development in the field of computational and statistical, quantitative and computational (gen)omics.
Figures

References
-
- Shipley M. Chronic widespread pain and fibromyalgia syndrome. Medicine 2018;46:252–5. 10.1016/j.mpmed.2018.01.009 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

