The structure of a virus-encoded nucleosome
- PMID: 33927388
- PMCID: PMC8370576
- DOI: 10.1038/s41594-021-00585-7
The structure of a virus-encoded nucleosome
Abstract
Certain large DNA viruses, including those in the Marseilleviridae family, encode histones. Here we show that fused histone pairs Hβ-Hα and Hδ-Hγ from Marseillevirus are structurally analogous to the eukaryotic histone pairs H2B-H2A and H4-H3. These viral histones form 'forced' heterodimers, and a heterotetramer of four such heterodimers assembles DNA to form structures virtually identical to canonical eukaryotic nucleosomes.
Figures

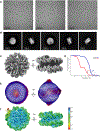
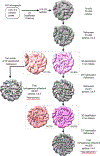






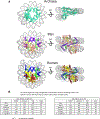
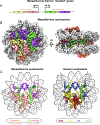


Comment in
-
A small nucleosome from a weird virus with a fat genome.Mol Cell. 2021 Sep 2;81(17):3447-3448. doi: 10.1016/j.molcel.2021.08.014. Mol Cell. 2021. PMID: 34478653
References
-
- Talbert PB, Meers MP & Henikoff S. Old cogs, new tricks: the evolution of gene expression in a chromatin context. Nat Rev Genet 20, 283–297 (2019). - PubMed
-
- Kornberg RD Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–71 (1974). - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF & Richmond TJ Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–60 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

