Molecular Programming of Drought-Challenged Trichoderma harzianum-Bioprimed Rice (Oryza sativa L.)
- PMID: 33927706
- PMCID: PMC8076752
- DOI: 10.3389/fmicb.2021.655165
Molecular Programming of Drought-Challenged Trichoderma harzianum-Bioprimed Rice (Oryza sativa L.)
Abstract
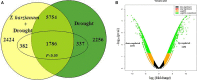
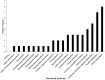
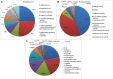
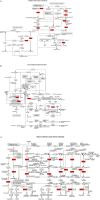
Trichoderma biopriming enhances rice growth in drought-stressed soils by triggering various plant metabolic pathways related to antioxidative defense, secondary metabolites, and hormonal upregulation. In the present study, transcriptomic analysis of rice cultivar IR64 bioprimed with Trichoderma harzianum under drought stress was carried out in comparison with drought-stressed samples using next-generation sequencing techniques. Out of the 2,506 significant (p < 0.05) differentially expressed genes (DEGs), 337 (15%) were exclusively expressed in drought-stressed plants, 382 (15%) were expressed in T. harzianum-treated drought-stressed plants, and 1,787 (70%) were commonly expressed. Furthermore, comparative analysis of upregulated and downregulated genes under stressed conditions showed that 1,053 genes (42%) were upregulated and 733 genes (29%) were downregulated in T. harzianum-treated drought-stressed rice plants. The genes exclusively expressed in T. harzianum-treated drought-stressed plants were mostly photosynthetic and antioxidative such as plastocyanin, small chain of Rubisco, PSI subunit Q, PSII subunit PSBY, osmoproteins, proline-rich protein, aquaporins, stress-enhanced proteins, and chaperonins. The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis states that the most enriched pathways were metabolic (38%) followed by pathways involved in the synthesis of secondary metabolites (25%), carbon metabolism (6%), phenyl propanoid (7%), and glutathione metabolism (3%). Some of the genes were selected for validation using real-time PCR which showed consistent expression as RNA-Seq data. Furthermore, to establish host-T. harzianum interaction, transcriptome analysis of Trichoderma was also carried out. The Gene Ontology (GO) analysis of T. harzianum transcriptome suggested that the annotated genes are functionally related to carbohydrate binding module, glycoside hydrolase, GMC oxidoreductase, and trehalase and were mainly upregulated, playing an important role in establishing the mycelia colonization of rice roots and its growth. Overall, it can be concluded that T. harzianum biopriming delays drought stress in rice cultivars by a multitude of molecular programming.
Keywords: DEGs; Trichoderma harzianum; drought; rice; transcriptome (RNA-seq).
Copyright © 2021 Bashyal, Parmar, Zaidi and Aggarwal.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Alwhibi M. S., Hashem A., Fathi E., Allah A., Alqarawi A. A., Soliman D. W. K., et al. (2017). Increased resistance of drought by Trichoderma harzianum fungal treatment correlates with increased secondary metabolites and proline content. J. Integ. Agric. 16 1751–1757. 10.1016/S2095-3119(17)61695-2 - DOI
-
- Azarmi R., Hajieghrari B., Giglou A. (2011). Effect of Trichoderma isolates on tomato seedling growth response and nutrient uptake. Afric. J. Biotechnol. 10 5850–5855. 10.5897/AJB10.1600 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

